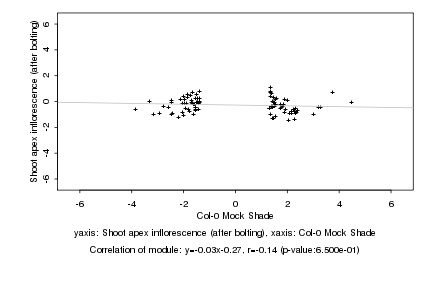
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col-0 Mock Shade |
Log2 signal ratio
Shoot apex inflorescence (after bolting) |
| 1 |
AT1G04180 |
AT1G04180
|
YUC9, YUCCA 9 |
4.467 |
-0.033 |
| 2 |
ATCG00950 |
ATCG00950
|
RRN23S.1, ribosomal RNA23S |
4.203 |
no data |
| 3 |
AT2G44910 |
AT2G44910
|
ATHB-4, ARABIDOPSIS THALIANA HOMEOBOX-LEUCINE ZIPPER PROTEIN 4, ATHB4, homeobox-leucine zipper protein 4, HB4, homeobox-leucine zipper protein 4 |
3.715 |
0.774 |
| 4 |
AT4G31380 |
AT4G31380
|
FLP1, FPF1-like protein 1 |
3.570 |
no data |
| 5 |
AT5G36150 |
AT5G36150
|
ATPEN3, putative pentacyclic triterpene synthase 3, PEN3, putative pentacyclic triterpene synthase 3 |
3.245 |
-0.454 |
| 6 |
AT2G38240 |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
3.192 |
-0.386 |
| 7 |
AT3G21330 |
AT3G21330
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
3.114 |
no data |
| 8 |
AT4G16780 |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
2.991 |
-0.965 |
| 9 |
AT4G37770 |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
2.393 |
-0.684 |
| 10 |
AT3G21320 |
AT3G21320
|
unknown |
2.332 |
no data |
| 11 |
AT5G43890 |
AT5G43890
|
SUPER1, SUPPRESSOR OF ER 1, YUC5, YUCCA5 |
2.325 |
-0.788 |
| 12 |
AT4G28720 |
AT4G28720
|
YUC8, YUCCA 8 |
2.313 |
-0.506 |
| 13 |
AT2G46970 |
AT2G46970
|
PIL1, phytochrome interacting factor 3-like 1 |
2.312 |
no data |
| 14 |
AT3G21720 |
AT3G21720
|
ICL, isocitrate lyase |
2.307 |
-0.855 |
| 15 |
AT3G03830 |
AT3G03830
|
SAUR28, SMALL AUXIN UP RNA 28 |
2.250 |
-1.389 |
| 16 |
AT4G32280 |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
2.245 |
-0.730 |
| 17 |
AT1G62420 |
AT1G62420
|
unknown |
2.224 |
no data |
| 18 |
AT5G12050 |
AT5G12050
|
unknown |
2.219 |
-0.615 |
| 19 |
AT3G51400 |
AT3G51400
|
unknown |
2.143 |
-0.737 |
| 20 |
AT1G15580 |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
2.137 |
-0.907 |
| 21 |
AT3G61920 |
AT3G61920
|
unknown |
2.124 |
-0.772 |
| 22 |
AT5G66580 |
AT5G66580
|
unknown |
2.104 |
no data |
| 23 |
AT5G18050 |
AT5G18050
|
SAUR22, SMALL AUXIN UP RNA 22 |
2.100 |
no data |
| 24 |
AT4G36700 |
AT4G36700
|
[RmlC-like cupins superfamily protein] |
2.079 |
-0.926 |
| 25 |
AT1G29490 |
AT1G29490
|
SAUR68, SMALL AUXIN UPREGULATED 68 |
2.031 |
-1.462 |
| 26 |
AT5G47370 |
AT5G47370
|
HAT2 |
1.971 |
0.153 |
| 27 |
AT5G18010 |
AT5G18010
|
SAUR19, small auxin up RNA 19 |
1.959 |
no data |
| 28 |
AT5G57640 |
AT5G57640
|
[GCK domain-containing protein] |
1.918 |
no data |
| 29 |
AT3G15170 |
AT3G15170
|
ANAC054, Arabidopsis NAC domain containing protein 54, ATNAC1, CUC1, CUP-SHAPED COTYLEDON1 |
1.892 |
-0.551 |
| 30 |
AT2G42870 |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
1.865 |
-0.775 |
| 31 |
ATCG00020 |
ATCG00020
|
PSBA, photosystem II reaction center protein A |
1.856 |
0.197 |
| 32 |
ATCG00350 |
ATCG00350
|
PSAA |
1.828 |
-0.222 |
| 33 |
AT4G34770 |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
1.810 |
-0.378 |
| 34 |
ATCG00490 |
ATCG00490
|
RBCL |
1.745 |
-0.398 |
| 35 |
AT1G02620 |
AT1G02620
|
[Ras-related small GTP-binding family protein] |
1.725 |
-0.521 |
| 36 |
AT4G13260 |
AT4G13260
|
AtYUC2, YUC2, YUCCA2 |
1.702 |
-0.199 |
| 37 |
AT3G50800 |
AT3G50800
|
unknown |
1.685 |
no data |
| 38 |
AT1G11735 |
AT1G11735
|
MIR171B, microRNA171B |
1.673 |
no data |
| 39 |
AT1G17345 |
AT1G17345
|
SAUR77, SMALL AUXIN UPREGULATED RNA 77 |
1.605 |
no data |
| 40 |
AT3G03820 |
AT3G03820
|
SAUR29, SMALL AUXIN UP RNA 29 |
1.592 |
no data |
| 41 |
ATCG00040 |
ATCG00040
|
MATK, maturase K |
1.547 |
0.301 |
| 42 |
AT3G23030 |
AT3G23030
|
IAA2, indole-3-acetic acid inducible 2 |
1.543 |
0.221 |
| 43 |
AT1G29450 |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
1.519 |
-1.104 |
| 44 |
AT5G46240 |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
1.519 |
-0.202 |
| 45 |
AT2G18010 |
AT2G18010
|
SAUR10, SMALL AUXIN UPREGULATED RNA 10 |
1.513 |
-0.226 |
| 46 |
AT1G50040 |
AT1G50040
|
unknown |
1.505 |
-0.041 |
| 47 |
AT3G55500 |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
1.482 |
-0.375 |
| 48 |
AT2G01021 |
AT2G01021
|
unknown |
1.477 |
no data |
| 49 |
AT3G03850 |
AT3G03850
|
SAUR26, SMALL AUXIN UP RNA 26 |
1.474 |
no data |
| 50 |
AT5G64750 |
AT5G64750
|
ABR1, ABA REPRESSOR1 |
1.474 |
no data |
| 51 |
AT5G65800 |
AT5G65800
|
ACS5, ACC synthase 5, ATACS5, CIN5, CYTOKININ-INSENSITIVE 5, ETO2, ETHYLENE OVERPRODUCER 2 |
1.449 |
-1.284 |
| 52 |
AT5G18060 |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
1.448 |
0.348 |
| 53 |
AT5G02760 |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
1.432 |
-0.001 |
| 54 |
AT1G22810 |
AT1G22810
|
[Integrase-type DNA-binding superfamily protein] |
1.420 |
-1.253 |
| 55 |
AT2G27690 |
AT2G27690
|
CYP94C1, cytochrome P450, family 94, subfamily C, polypeptide 1 |
1.414 |
-0.377 |
| 56 |
AT4G17460 |
AT4G17460
|
HAT1, JAB, JAIBA |
1.412 |
-0.393 |
| 57 |
AT5G39860 |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
1.399 |
0.033 |
| 58 |
AT2G14245 |
AT2G14245
|
[copia-like retrotransposon family, has a 2.2e-28 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
1.398 |
no data |
| 59 |
AT3G41768 |
AT3G41768
|
[18SrRNA] |
1.387 |
no data |
| 60 |
ATMG00020 |
ATMG00020
|
RRN26, ribosomal RNA26S |
1.381 |
no data |
| 61 |
AT3G15540 |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
1.375 |
-0.368 |
| 62 |
ATCG00280 |
ATCG00280
|
PSBC, photosystem II reaction center protein C |
1.364 |
0.686 |
| 63 |
AT1G10550 |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
1.347 |
0.429 |
| 64 |
AT3G60390 |
AT3G60390
|
HAT3, homeobox-leucine zipper protein 3 |
1.345 |
0.794 |
| 65 |
AT4G18170 |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
1.339 |
-0.976 |
| 66 |
ATCG00340 |
ATCG00340
|
PSAB |
1.337 |
1.134 |
| 67 |
AT2G26710 |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
1.337 |
0.773 |
| 68 |
AT1G72620 |
AT1G72620
|
[alpha/beta-Hydrolases superfamily protein] |
1.331 |
no data |
| 69 |
AT5G18030 |
AT5G18030
|
[SAUR-like auxin-responsive protein family ] |
1.321 |
no data |
| 70 |
AT1G73550 |
AT1G73550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.313 |
no data |
| 71 |
AT5G62280 |
AT5G62280
|
unknown |
1.309 |
-0.499 |
| 72 |
AT2G01300 |
AT2G01300
|
unknown |
1.296 |
-0.528 |
| 73 |
AT2G41240 |
AT2G41240
|
BHLH100, basic helix-loop-helix protein 100 |
1.293 |
no data |
| 74 |
ATCG00330 |
ATCG00330
|
RPS14, chloroplast ribosomal protein S14 |
1.283 |
-0.528 |
| 75 |
AT1G58983 |
AT1G58983
|
[Ribosomal protein S5 family protein] |
-6.350 |
no data |
| 76 |
AT4G34410 |
AT4G34410
|
RRTF1, redox responsive transcription factor 1 |
-3.892 |
-0.599 |
| 77 |
AT2G15020 |
AT2G15020
|
unknown |
-3.337 |
0.021 |
| 78 |
AT1G05880 |
AT1G05880
|
ARI12, ARIADNE 12, ATARI12, ARABIDOPSIS ARIADNE 12 |
-3.175 |
-0.932 |
| 79 |
AT3G61840 |
AT3G61840
|
unknown |
-2.946 |
-0.852 |
| 80 |
AT4G15248 |
AT4G15248
|
BBX30, B-box domain protein 30 |
-2.792 |
no data |
| 81 |
AT3G21890 |
AT3G21890
|
BBX31, B-box domain protein 31 |
-2.776 |
-0.379 |
| 82 |
AT3G21150 |
AT3G21150
|
B-box 32 |
-2.694 |
no data |
| 83 |
AT2G27420 |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-2.607 |
-0.412 |
| 84 |
AT4G13395 |
AT4G13395
|
DVL10, DEVIL 10, RTFL12, ROTUNDIFOLIA like 12 |
-2.539 |
no data |
| 85 |
AT4G25480 |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
-2.492 |
-0.992 |
| 86 |
AT5G45630 |
AT5G45630
|
unknown |
-2.492 |
0.106 |
| 87 |
AT3G22231 |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-2.482 |
-0.065 |
| 88 |
AT2G37720 |
AT2G37720
|
TBL15, TRICHOME BIREFRINGENCE-LIKE 15 |
-2.445 |
-0.858 |
| 89 |
AT4G34550 |
AT4G34550
|
unknown |
-2.316 |
no data |
| 90 |
AT4G23515 |
AT4G23515
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-2.255 |
no data |
| 91 |
AT4G26950 |
AT4G26950
|
unknown |
-2.216 |
-1.183 |
| 92 |
AT3G27250 |
AT3G27250
|
unknown |
-2.189 |
no data |
| 93 |
AT3G44450 |
AT3G44450
|
unknown |
-2.134 |
0.171 |
| 94 |
AT1G72910 |
AT1G72910
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
-2.113 |
no data |
| 95 |
AT2G36750 |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
-2.078 |
-0.821 |
| 96 |
AT5G44420 |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
-2.076 |
-0.110 |
| 97 |
AT4G04990 |
AT4G04990
|
unknown |
-2.031 |
-1.069 |
| 98 |
AT2G29090 |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
-2.015 |
0.436 |
| 99 |
AT1G26390 |
AT1G26390
|
[FAD-binding Berberine family protein] |
-1.982 |
-0.140 |
| 100 |
AT3G21760 |
AT3G21760
|
HYR1, HYPOSTATIN RESISTANCE 1 |
-1.968 |
0.223 |
| 101 |
AT1G09240 |
AT1G09240
|
ATNAS3, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 3, NAS3, nicotianamine synthase 3 |
-1.934 |
-0.521 |
| 102 |
AT1G70260 |
AT1G70260
|
UMAMIT36, Usually multiple acids move in and out Transporters 36 |
-1.928 |
no data |
| 103 |
AT4G27657 |
AT4G27657
|
unknown |
-1.899 |
-0.116 |
| 104 |
AT5G50800 |
AT5G50800
|
AtSWEET13, SWEET13 |
-1.872 |
0.324 |
| 105 |
AT5G49740 |
AT5G49740
|
ATFRO7, FERRIC REDUCTION OXIDASE 7, FRO7, ferric reduction oxidase 7 |
-1.854 |
no data |
| 106 |
AT3G19270 |
AT3G19270
|
CYP707A4, cytochrome P450, family 707, subfamily A, polypeptide 4 |
-1.852 |
0.577 |
| 107 |
AT1G72920 |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-1.844 |
-0.584 |
| 108 |
AT5G42530 |
AT5G42530
|
unknown |
-1.837 |
no data |
| 109 |
AT4G27360 |
AT4G27360
|
[Dynein light chain type 1 family protein] |
-1.809 |
no data |
| 110 |
AT2G46790 |
AT2G46790
|
APRR9, Arabidopsis pseudo-response regulator 9, PRR9, pseudo-response regulator 9, TL1, TOC1-LIKE PROTEIN 1 |
-1.805 |
no data |
| 111 |
AT3G09450 |
AT3G09450
|
unknown |
-1.787 |
no data |
| 112 |
AT5G46830 |
AT5G46830
|
ATNIG1, NACL-INDUCIBLE GENE 1, NIG1, NACL-inducible gene 1 |
-1.777 |
-0.767 |
| 113 |
AT2G36800 |
AT2G36800
|
DOGT1, don-glucosyltransferase 1, UGT73C5, UDP-GLUCOSYL TRANSFERASE 73C5 |
-1.757 |
no data |
| 114 |
AT3G49160 |
AT3G49160
|
[pyruvate kinase family protein] |
-1.756 |
0.540 |
| 115 |
AT4G12320 |
AT4G12320
|
CYP706A6, cytochrome P450, family 706, subfamily A, polypeptide 6 |
-1.743 |
no data |
| 116 |
AT3G15310 |
AT3G15310
|
unknown |
-1.733 |
0.093 |
| 117 |
AT2G31230 |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
-1.728 |
-0.027 |
| 118 |
AT5G24155 |
AT5G24155
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
-1.710 |
no data |
| 119 |
AT4G29110 |
AT4G29110
|
unknown |
-1.678 |
no data |
| 120 |
AT1G55960 |
AT1G55960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-1.678 |
-0.079 |
| 121 |
AT4G33040 |
AT4G33040
|
[Thioredoxin superfamily protein] |
-1.671 |
-0.091 |
| 122 |
AT2G21320 |
AT2G21320
|
BBX18, B-box domain protein 18 |
-1.656 |
0.751 |
| 123 |
AT4G16880 |
AT4G16880
|
[Leucine-rich repeat (LRR) family protein] |
-1.656 |
no data |
| 124 |
AT1G76530 |
AT1G76530
|
[Auxin efflux carrier family protein] |
-1.652 |
-0.996 |
| 125 |
AT1G66540 |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
-1.645 |
-0.182 |
| 126 |
AT5G45850 |
AT5G45850
|
unknown |
-1.608 |
-0.221 |
| 127 |
AT3G23170 |
AT3G23170
|
unknown |
-1.564 |
0.262 |
| 128 |
AT3G04140 |
AT3G04140
|
[Ankyrin repeat family protein] |
-1.562 |
no data |
| 129 |
AT5G17350 |
AT5G17350
|
unknown |
-1.557 |
-0.646 |
| 130 |
AT5G66740 |
AT5G66740
|
unknown |
-1.556 |
-0.591 |
| 131 |
AT4G26530 |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
-1.544 |
-0.399 |
| 132 |
AT3G52740 |
AT3G52740
|
unknown |
-1.543 |
-0.399 |
| 133 |
AT5G54130 |
AT5G54130
|
[Calcium-binding endonuclease/exonuclease/phosphatase family] |
-1.538 |
-0.576 |
| 134 |
AT5G54470 |
AT5G54470
|
BBX29, B-box domain protein 29 |
-1.510 |
0.561 |
| 135 |
AT1G26410 |
AT1G26410
|
[FAD-binding Berberine family protein] |
-1.501 |
-0.063 |
| 136 |
AT3G47430 |
AT3G47430
|
PEX11B, peroxin 11B |
-1.496 |
-0.014 |
| 137 |
AT1G78995 |
AT1G78995
|
unknown |
-1.468 |
0.247 |
| 138 |
AT5G17300 |
AT5G17300
|
RVE1, REVEILLE 1 |
-1.456 |
-0.599 |
| 139 |
AT1G48100 |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-1.453 |
-0.111 |
| 140 |
AT1G76955 |
AT1G76955
|
[Expressed protein] |
-1.440 |
no data |
| 141 |
AT4G11280 |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
-1.439 |
-0.138 |
| 142 |
AT4G27652 |
AT4G27652
|
unknown |
-1.423 |
0.245 |
| 143 |
AT5G24660 |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-1.422 |
0.006 |
| 144 |
AT1G18330 |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
-1.405 |
0.784 |
| 145 |
AT5G59820 |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
-1.402 |
-0.012 |
| 146 |
AT2G29170 |
AT2G29170
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-1.397 |
no data |
| 147 |
AT1G24147 |
AT1G24147
|
unknown |
-1.387 |
no data |