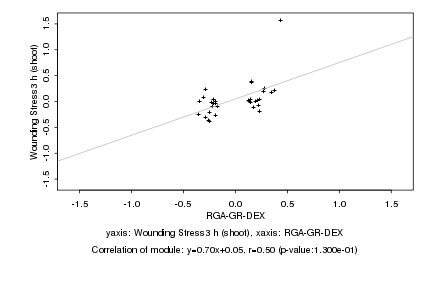
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
RGA-GR-DEX |
Log2 signal ratio
Wounding Stress3 h (shoot) |
| 1 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.425 |
1.585 |
| 2 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.367 |
0.217 |
| 3 |
255733_at |
AT1G25400
|
unknown |
0.346 |
0.188 |
| 4 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
0.277 |
0.252 |
| 5 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.261 |
0.194 |
| 6 |
251356_at |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
0.230 |
-0.179 |
| 7 |
255057_at |
AT4G09840
|
unknown |
0.225 |
0.050 |
| 8 |
267220_at |
AT2G02500
|
ATMEPCT, ISPD, MCT, 2-C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLTRANSFERASE |
0.221 |
-0.059 |
| 9 |
250223_at |
AT5G14070
|
ROXY2 |
0.207 |
0.032 |
| 10 |
247490_at |
AT5G61850
|
LFY, LEAFY, LFY3, LEAFY 3 |
0.192 |
0.014 |
| 11 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.167 |
-0.103 |
| 12 |
262801_at |
AT1G21010
|
unknown |
0.150 |
0.368 |
| 13 |
245731_at |
AT1G73500
|
ATMKK9, MKK9, MAP kinase kinase 9 |
0.147 |
0.398 |
| 14 |
247299_at |
AT5G63900
|
[Acyl-CoA N-acyltransferase with RING/FYVE/PHD-type zinc finger domain] |
0.141 |
-0.015 |
| 15 |
250790_at |
AT5G05640
|
[nucleoprotein-related] |
0.137 |
0.058 |
| 16 |
245098_at |
AT2G40940
|
ERS, ETHYLENE RESPONSE SENSOR, ERS1, ethylene response sensor 1 |
0.134 |
0.008 |
| 17 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.123 |
0.028 |
| 18 |
252591_at |
AT3G45600
|
TET3, tetraspanin3 |
-0.358 |
-0.249 |
| 19 |
247447_at |
AT5G62730
|
[Major facilitator superfamily protein] |
-0.350 |
0.010 |
| 20 |
250936_at |
AT5G03120
|
unknown |
-0.312 |
0.087 |
| 21 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
-0.296 |
0.247 |
| 22 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.289 |
-0.295 |
| 23 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
-0.261 |
-0.348 |
| 24 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.258 |
-0.196 |
| 25 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
-0.252 |
-0.379 |
| 26 |
254762_at |
AT4G13230
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.234 |
-0.002 |
| 27 |
248924_at |
AT5G45960
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.221 |
-0.077 |
| 28 |
263238_at |
AT2G16580
|
SAUR8, SMALL AUXIN UPREGULATED RNA 8 |
-0.219 |
-0.026 |
| 29 |
251293_at |
AT3G61930
|
unknown |
-0.213 |
0.043 |
| 30 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.201 |
-0.037 |
| 31 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.194 |
-0.263 |
| 32 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
-0.194 |
0.016 |
| 33 |
250933_at |
AT5G03170
|
ATFLA11, ARABIDOPSIS FASCICLIN-LIKE ARABINOGALACTAN-PROTEIN 11, FLA11, FASCICLIN-like arabinogalactan-protein 11 |
-0.178 |
-0.082 |