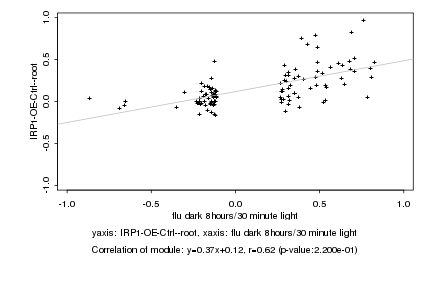
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
flu dark 8hours/30 minute light |
Log2 signal ratio
IRP1-OE-Ctrl--root |
| 1 |
248799_at |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
0.820 |
0.477 |
| 2 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.804 |
0.295 |
| 3 |
266010_at |
AT2G37430
|
ZAT11, zinc finger of Arabidopsis thaliana 11 |
0.798 |
0.395 |
| 4 |
258653_at |
AT3G09870
|
SAUR48, SMALL AUXIN UPREGULATED RNA 48 |
0.780 |
0.056 |
| 5 |
256442_at |
AT3G10930
|
unknown |
0.756 |
0.978 |
| 6 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.706 |
0.520 |
| 7 |
256306_at |
AT1G30370
|
DLAH, DAD1-like acylhydrolase |
0.704 |
0.359 |
| 8 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.684 |
0.824 |
| 9 |
263182_at |
AT1G05575
|
unknown |
0.678 |
0.387 |
| 10 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
0.677 |
0.480 |
| 11 |
247047_at |
AT5G66650
|
unknown |
0.642 |
0.211 |
| 12 |
245755_at |
AT1G35210
|
unknown |
0.635 |
0.439 |
| 13 |
261476_at |
AT1G14480
|
[Ankyrin repeat family protein] |
0.624 |
0.286 |
| 14 |
246584_at |
AT5G14730
|
unknown |
0.607 |
0.463 |
| 15 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.561 |
0.417 |
| 16 |
253044_at |
AT4G37290
|
unknown |
0.537 |
0.174 |
| 17 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
0.534 |
0.023 |
| 18 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.534 |
0.200 |
| 19 |
262731_at |
AT1G16420
|
ATMC8, ARABIDOPSIS THALIANA METACASPASE 8, AtMCP2e, metacaspase 2e, MC8, metacaspase 8, MCP2e, metacaspase 2e |
0.519 |
-0.002 |
| 20 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
0.516 |
0.344 |
| 21 |
253643_at |
AT4G29780
|
unknown |
0.483 |
0.650 |
| 22 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.483 |
0.359 |
| 23 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.482 |
0.470 |
| 24 |
248726_at |
AT5G47960
|
ATRABA4C, RAB GTPase homolog A4C, RABA4C, RAB GTPase homolog A4C, SMG1, SMALL MOLECULAR WEIGHT G-PROTEIN 1 |
0.477 |
0.198 |
| 25 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
0.473 |
0.795 |
| 26 |
265725_at |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.470 |
0.298 |
| 27 |
259979_at |
AT1G76600
|
unknown |
0.443 |
0.159 |
| 28 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.424 |
0.681 |
| 29 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.402 |
0.270 |
| 30 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.390 |
0.763 |
| 31 |
249770_at |
AT5G24110
|
ATWRKY30, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 30, WRKY30, WRKY DNA-binding protein 30 |
0.377 |
-0.066 |
| 32 |
267623_at |
AT2G39650
|
unknown |
0.373 |
0.308 |
| 33 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.370 |
0.056 |
| 34 |
252193_at |
AT3G50060
|
MYB77, myb domain protein 77 |
0.352 |
0.387 |
| 35 |
260037_at |
AT1G68840
|
AtRAV2, EDF2, ETHYLENE RESPONSE DNA BINDING FACTOR 2, RAP2.8, RELATED TO AP2 8, RAV2, related to ABI3/VP1 2, TEM2, TEMPRANILLO 2 |
0.350 |
0.098 |
| 36 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.350 |
0.280 |
| 37 |
262383_at |
AT1G72940
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.348 |
0.103 |
| 38 |
247351_at |
AT5G63790
|
ANAC102, NAC domain containing protein 102, NAC102, NAC domain containing protein 102 |
0.326 |
0.196 |
| 39 |
253281_at |
AT4G34138
|
UGT73B1, UDP-glucosyl transferase 73B1 |
0.317 |
0.023 |
| 40 |
248392_at |
AT5G52050
|
[MATE efflux family protein] |
0.315 |
0.358 |
| 41 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.315 |
0.321 |
| 42 |
248870_at |
AT5G46710
|
[PLATZ transcription factor family protein] |
0.313 |
-0.029 |
| 43 |
247753_at |
AT5G59070
|
[UDP-Glycosyltransferase superfamily protein] |
0.311 |
0.067 |
| 44 |
245731_at |
AT1G73500
|
ATMKK9, MKK9, MAP kinase kinase 9 |
0.310 |
0.159 |
| 45 |
252474_at |
AT3G46620
|
unknown |
0.299 |
0.244 |
| 46 |
265184_at |
AT1G23710
|
unknown |
0.294 |
0.321 |
| 47 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
0.292 |
-0.112 |
| 48 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
0.290 |
0.260 |
| 49 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
0.287 |
0.438 |
| 50 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
0.280 |
0.028 |
| 51 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.277 |
0.151 |
| 52 |
260477_at |
AT1G11050
|
[Protein kinase superfamily protein] |
0.275 |
0.130 |
| 53 |
258682_at |
AT3G08720
|
ATPK19, Arabidopsis thaliana protein kinase 19, ATPK2, ARABIDOPSIS THALIANA PROTEIN KINASE 2, ATS6K2, ARABIDOPSIS THALIANA SERINE/THREONINE PROTEIN KINASE 2, S6K2, serine/threonine protein kinase 2 |
0.271 |
-0.002 |
| 54 |
262384_at |
AT1G72950
|
[Disease resistance protein (TIR-NBS class)] |
0.270 |
0.128 |
| 55 |
255733_at |
AT1G25400
|
unknown |
0.270 |
0.032 |
| 56 |
263379_at |
AT2G40140
|
ATSZF2, CZF1, SZF2, (SALT-INDUCIBLE ZINC FINGER 2, ZFAR1 |
0.267 |
0.222 |
| 57 |
265441_at |
AT2G20870
|
[cell wall protein precursor, putative] |
0.267 |
0.050 |
| 58 |
258027_at |
AT3G19515
|
unknown |
-0.869 |
0.043 |
| 59 |
258424_at |
AT3G16750
|
unknown |
-0.693 |
-0.077 |
| 60 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
-0.659 |
-0.040 |
| 61 |
258113_at |
AT3G14650
|
CYP72A11, cytochrome P450, family 72, subfamily A, polypeptide 11 |
-0.657 |
0.004 |
| 62 |
257067_at |
AT3G18270
|
pseudogene |
-0.356 |
-0.061 |
| 63 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
-0.307 |
0.112 |
| 64 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
-0.237 |
0.011 |
| 65 |
258050_at |
AT3G16200
|
unknown |
-0.230 |
-0.022 |
| 66 |
258633_at |
AT3G07990
|
SCPL27, serine carboxypeptidase-like 27 |
-0.221 |
-0.019 |
| 67 |
251058_at |
AT5G01790
|
unknown |
-0.219 |
0.002 |
| 68 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
-0.218 |
-0.144 |
| 69 |
263829_at |
AT2G40435
|
unknown |
-0.217 |
0.044 |
| 70 |
256249_at |
AT3G11270
|
MEE34, maternal effect embryo arrest 34 |
-0.209 |
-0.027 |
| 71 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-0.206 |
0.126 |
| 72 |
248918_at |
AT5G45890
|
AtSAG12, SAG12, senescence-associated gene 12 |
-0.205 |
-0.018 |
| 73 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-0.203 |
0.221 |
| 74 |
246159_at |
AT5G20935
|
CRR42, CHLORORESPIRATORY REDUCTION 42 |
-0.190 |
0.066 |
| 75 |
263580_at |
AT2G17140
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.189 |
0.181 |
| 76 |
247064_at |
AT5G66890
|
[Leucine-rich repeat (LRR) family protein] |
-0.184 |
0.002 |
| 77 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
-0.180 |
-0.039 |
| 78 |
259102_at |
AT3G11660
|
NHL1, NDR1/HIN1-like 1 |
-0.178 |
0.085 |
| 79 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
-0.177 |
0.088 |
| 80 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
-0.168 |
0.180 |
| 81 |
257729_at |
AT3G18520
|
ATHDA15, HDA15, histone deacetylase 15 |
-0.167 |
-0.098 |
| 82 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
-0.163 |
0.045 |
| 83 |
255773_at |
AT1G18590
|
ATSOT17, SULFOTRANSFERASE 17, ATST5C, ARABIDOPSIS SULFOTRANSFERASE 5C, SOT17, sulfotransferase 17 |
-0.158 |
0.176 |
| 84 |
249166_at |
AT5G42840
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.153 |
-0.014 |
| 85 |
262156_at |
AT1G52680
|
[late embryogenesis abundant protein-related / LEA protein-related] |
-0.152 |
0.146 |
| 86 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
-0.151 |
0.155 |
| 87 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
-0.149 |
-0.025 |
| 88 |
257730_at |
AT3G18420
|
[Protein prenylyltransferase superfamily protein] |
-0.148 |
0.110 |
| 89 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
-0.147 |
0.282 |
| 90 |
250425_at |
AT5G10530
|
LecRK-IX.1, L-type lectin receptor kinase IX.1 |
-0.147 |
0.003 |
| 91 |
246797_at |
AT5G26790
|
unknown |
-0.145 |
-0.124 |
| 92 |
256568_at |
AT3G19520
|
unknown |
-0.143 |
-0.004 |
| 93 |
257271_at |
AT3G28007
|
AtSWEET4, SWEET4 |
-0.142 |
0.072 |
| 94 |
264880_at |
AT1G61210
|
DWA3, DWD hypersensitive to ABA 3 |
-0.140 |
0.165 |
| 95 |
256673_at |
AT3G52370
|
FLA15, FASCICLIN-like arabinogalactan protein 15 precursor |
-0.136 |
0.057 |
| 96 |
249788_at |
AT5G24330
|
ATXR6, ARABIDOPSIS TRITHORAX-RELATED PROTEIN 6, SDG34, SET DOMAIN PROTEIN 34 |
-0.133 |
-0.043 |
| 97 |
254512_at |
AT4G20230
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.131 |
-0.029 |
| 98 |
253165_at |
AT4G35320
|
unknown |
-0.131 |
0.091 |
| 99 |
257108_x_at |
AT3G30490
|
unknown |
-0.131 |
0.066 |
| 100 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.130 |
0.088 |
| 101 |
259387_at |
AT1G13370
|
[Histone superfamily protein] |
-0.129 |
-0.033 |
| 102 |
253021_at |
AT4G38050
|
[Xanthine/uracil permease family protein] |
-0.128 |
0.009 |
| 103 |
248605_at |
AT5G49410
|
unknown |
-0.128 |
0.010 |
| 104 |
245897_at |
AT5G09400
|
KUP7, K+ uptake permease 7 |
-0.127 |
0.482 |
| 105 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
-0.125 |
-0.146 |
| 106 |
255896_at |
AT1G17800
|
AtENODL22, ENODL22, early nodulin-like protein 22 |
-0.123 |
0.094 |
| 107 |
258396_at |
AT3G15460
|
[Ribosomal RNA processing Brix domain protein] |
-0.122 |
0.067 |
| 108 |
261380_at |
AT1G05400
|
unknown |
-0.121 |
0.133 |
| 109 |
262137_at |
AT1G77920
|
TGA7, TGACG sequence-specific binding protein 7 |
-0.119 |
0.126 |
| 110 |
246516_at |
AT5G15740
|
[O-fucosyltransferase family protein] |
-0.119 |
0.001 |
| 111 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
-0.119 |
-0.158 |
| 112 |
249151_at |
AT5G43360
|
ATPT4, PHT1;3, phosphate transporter 1;3, PHT3, PHOSPHATE TRANSPORTER 3 |
-0.117 |
0.061 |
| 113 |
259655_at |
AT1G55210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.116 |
0.121 |
| 114 |
254737_at |
AT4G13840
|
CER26, ECERIFERUM 26 |
-0.116 |
0.053 |