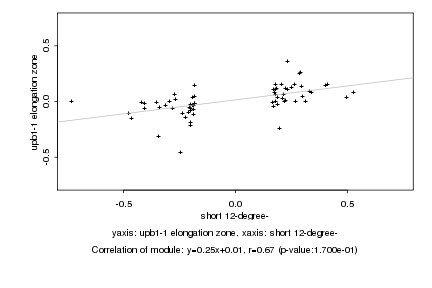
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
short 12-degree- |
Log2 signal ratio
upb1-1 elongation zone |
| 1 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.525 |
0.082 |
| 2 |
266532_at |
AT2G16890
|
[UDP-Glycosyltransferase superfamily protein] |
0.494 |
0.042 |
| 3 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
0.409 |
0.155 |
| 4 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
0.399 |
0.148 |
| 5 |
264261_at |
AT1G09240
|
ATNAS3, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 3, NAS3, nicotianamine synthase 3 |
0.339 |
0.085 |
| 6 |
249677_at |
AT5G35970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.327 |
0.096 |
| 7 |
251753_at |
AT3G55760
|
unknown |
0.313 |
0.007 |
| 8 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.299 |
0.054 |
| 9 |
245427_at |
AT4G17550
|
AtG3Pp4, glycerol-3-phosphate permease 4, G3Pp4, glycerol-3-phosphate permease 4 |
0.295 |
0.139 |
| 10 |
251827_at |
AT3G55120
|
A11, AtCHI, CFI, CHALCONE FLAVANONE ISOMERASE, CHI, chalcone isomerase, TT5, TRANSPARENT TESTA 5 |
0.289 |
0.263 |
| 11 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.283 |
0.260 |
| 12 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.267 |
0.008 |
| 13 |
246468_at |
AT5G17050
|
UGT78D2, UDP-glucosyl transferase 78D2 |
0.261 |
0.157 |
| 14 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
0.250 |
0.127 |
| 15 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
0.232 |
0.366 |
| 16 |
257237_at |
AT3G14890
|
[phosphoesterase] |
0.231 |
0.108 |
| 17 |
253841_at |
AT4G27830
|
AtBGLU10, BGLU10, beta glucosidase 10 |
0.221 |
0.017 |
| 18 |
261651_at |
AT1G27760
|
ATSAT32, SALT-TOLERANCE 32, SAT32, SALT-TOLERANCE 32 |
0.220 |
0.119 |
| 19 |
250151_at |
AT5G14570
|
ATNRT2.7, high affinity nitrate transporter 2.7, NRT2.7, high affinity nitrate transporter 2.7 |
0.219 |
0.005 |
| 20 |
264859_at |
AT1G24280
|
G6PD3, glucose-6-phosphate dehydrogenase 3 |
0.212 |
0.069 |
| 21 |
245200_at |
AT1G67850
|
unknown |
0.206 |
0.030 |
| 22 |
253252_at |
AT4G34740
|
ASE2, GLN phosphoribosyl pyrophosphate amidotransferase 2, ATASE2, GLN phosphoribosyl pyrophosphate amidotransferase 2, ATPURF2, CIA1, CHLOROPLAST IMPORT APPARATUS 1, DOV1, DIFFERENTIAL DEVELOPMENT OF VASCULAR ASSOCIATED CELLS |
0.204 |
0.159 |
| 23 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.196 |
-0.238 |
| 24 |
249038_at |
AT5G44280
|
ATRING1A, ARABIDOPSIS THALIANA RING 1A, RING1A, RING 1A |
0.186 |
-0.025 |
| 25 |
262881_at |
AT1G64890
|
[Major facilitator superfamily protein] |
0.186 |
0.044 |
| 26 |
263221_at |
AT1G30620
|
HSR8, HIGH SUGAR RESPONSE8, MUR4, MURUS 4, UXE1, UDP-D-XYLOSE 4-EPIMERASE 1 |
0.181 |
0.118 |
| 27 |
257517_at |
AT3G16330
|
unknown |
0.178 |
0.009 |
| 28 |
261818_at |
AT1G11390
|
[Protein kinase superfamily protein] |
0.178 |
0.068 |
| 29 |
265290_at |
AT2G22590
|
[UDP-Glycosyltransferase superfamily protein] |
0.175 |
0.158 |
| 30 |
249011_at |
AT5G44670
|
GALS2, galactan synthase 2 |
0.175 |
0.108 |
| 31 |
247025_at |
AT5G67030
|
ABA1, ABA DEFICIENT 1, ATABA1, ARABIDOPSIS THALIANA ABA DEFICIENT 1, ATZEP, ARABIDOPSIS THALIANA ZEAXANTHIN EPOXIDASE, IBS3, IMPAIRED IN BABA-INDUCED STERILITY 3, LOS6, LOW EXPRESSION OF OSMOTIC STRESS-RESPONSIVE GENES 6, NPQ2, NON-PHOTOCHEMICAL QUENCHING 2, ZEP, ZEAXANTHIN EPOXIDASE |
0.171 |
0.089 |
| 32 |
252956_at |
AT4G38580
|
ATFP6, farnesylated protein 6, FP6, farnesylated protein 6, HIPP26, HEAVY METAL ASSOCIATED ISOPRENYLATED PLANT PROTEIN 26 |
0.169 |
-0.042 |
| 33 |
249148_at |
AT5G43260
|
[chaperone protein dnaJ-related] |
0.166 |
0.116 |
| 34 |
261116_at |
AT1G75370
|
[Sec14p-like phosphatidylinositol transfer family protein] |
0.162 |
-0.003 |
| 35 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-0.736 |
0.001 |
| 36 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.481 |
-0.099 |
| 37 |
259104_at |
AT3G02170
|
LNG2, LONGIFOLIA2, TRM1, TON1 Recruiting Motif 1 |
-0.465 |
-0.148 |
| 38 |
260431_at |
AT1G68190
|
BBX27, B-box domain protein 27 |
-0.422 |
-0.003 |
| 39 |
263106_at |
AT2G05160
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
-0.409 |
-0.014 |
| 40 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.407 |
-0.061 |
| 41 |
252433_at |
AT3G47560
|
[alpha/beta-Hydrolases superfamily protein] |
-0.354 |
-0.003 |
| 42 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.344 |
-0.308 |
| 43 |
257615_at |
AT3G26510
|
[Octicosapeptide/Phox/Bem1p family protein] |
-0.342 |
-0.051 |
| 44 |
266106_at |
AT2G45170
|
ATATG8E, AUTOPHAGY 8E, ATG8E, AUTOPHAGY 8E |
-0.313 |
-0.027 |
| 45 |
260317_at |
AT1G63800
|
UBC5, ubiquitin-conjugating enzyme 5 |
-0.296 |
0.009 |
| 46 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
-0.282 |
-0.054 |
| 47 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
-0.275 |
0.070 |
| 48 |
247540_at |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
-0.270 |
0.026 |
| 49 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.248 |
-0.456 |
| 50 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
-0.237 |
-0.105 |
| 51 |
266265_at |
AT2G29340
|
[NAD-dependent epimerase/dehydratase family protein] |
-0.225 |
-0.139 |
| 52 |
261294_at |
AT1G48430
|
[Dihydroxyacetone kinase] |
-0.211 |
-0.098 |
| 53 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.209 |
-0.047 |
| 54 |
248398_at |
AT5G51970
|
[GroES-like zinc-binding alcohol dehydrogenase family protein] |
-0.205 |
-0.024 |
| 55 |
259017_at |
AT3G07310
|
unknown |
-0.205 |
-0.058 |
| 56 |
257772_at |
AT3G23080
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.204 |
-0.213 |
| 57 |
263118_at |
AT1G03090
|
MCCA |
-0.203 |
-0.188 |
| 58 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
-0.201 |
-0.072 |
| 59 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.196 |
-0.027 |
| 60 |
246487_at |
AT5G16030
|
unknown |
-0.192 |
0.039 |
| 61 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.191 |
-0.065 |
| 62 |
248795_at |
AT5G47390
|
MYBH, MYB hypocotyl elongation-related |
-0.189 |
-0.031 |
| 63 |
256455_at |
AT1G75190
|
unknown |
-0.188 |
-0.112 |
| 64 |
245362_at |
AT4G17460
|
HAT1, JAB, JAIBA |
-0.187 |
-0.011 |
| 65 |
247284_at |
AT5G64410
|
ATOPT4, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 4, OPT4, oligopeptide transporter 4 |
-0.187 |
0.149 |
| 66 |
257745_at |
AT3G29240
|
unknown |
-0.186 |
0.050 |
| 67 |
258935_at |
AT3G10120
|
unknown |
-0.184 |
-0.010 |