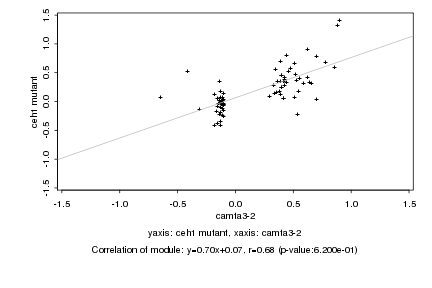
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
camta3-2 |
Log2 signal ratio
ceh1 mutant |
| 1 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.892 |
1.423 |
| 2 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.880 |
1.325 |
| 3 |
259385_at |
AT1G13470
|
unknown |
0.849 |
0.599 |
| 4 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.774 |
0.691 |
| 5 |
251673_at |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
0.694 |
0.041 |
| 6 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
0.693 |
0.796 |
| 7 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
0.652 |
0.326 |
| 8 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
0.634 |
0.339 |
| 9 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.621 |
0.908 |
| 10 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
0.619 |
0.418 |
| 11 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.582 |
0.317 |
| 12 |
263852_at |
AT2G04450
|
ATNUDT6, nudix hydrolase homolog 6, ATNUDX6, Arabidopsis thaliana nucleoside diphosphate linked to some moiety X 6, NUDT6, nudix hydrolase homolog 6, NUDX6, nucleoside diphosphates linked to some moiety X 6 |
0.550 |
0.401 |
| 13 |
255406_at |
AT4G03450
|
[Ankyrin repeat family protein] |
0.541 |
0.183 |
| 14 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.534 |
-0.215 |
| 15 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.521 |
0.370 |
| 16 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
0.512 |
0.480 |
| 17 |
247602_at |
AT5G60900
|
RLK1, receptor-like protein kinase 1 |
0.507 |
0.664 |
| 18 |
265658_at |
AT2G13810
|
ALD1, AGD2-like defense response protein 1, AtALD1, EDTS5, eds two suppressor 5 |
0.505 |
0.073 |
| 19 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
0.472 |
0.575 |
| 20 |
254500_at |
AT4G20110
|
BP80-3;1, binding protein of 80 kDa 3;1, VSR3;1, VACUOLAR SORTING RECEPTOR 3;1, VSR7, VACUOLAR SORTING RECEPTOR 7 |
0.450 |
0.525 |
| 21 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
0.437 |
0.801 |
| 22 |
254242_at |
AT4G23200
|
CRK12, cysteine-rich RLK (RECEPTOR-like protein kinase) 12 |
0.434 |
0.337 |
| 23 |
265161_at |
AT1G30900
|
BP80-3;3, binding protein of 80 kDa 3;3, VSR3;3, VACUOLAR SORTING RECEPTOR 3;3, VSR6, VACUOLAR SORTING RECEPTOR 6 |
0.422 |
0.417 |
| 24 |
249485_at |
AT5G39020
|
[Malectin/receptor-like protein kinase family protein] |
0.418 |
0.291 |
| 25 |
263182_at |
AT1G05575
|
unknown |
0.416 |
0.390 |
| 26 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.412 |
0.364 |
| 27 |
249918_at |
AT5G19240
|
[Glycoprotein membrane precursor GPI-anchored] |
0.410 |
0.060 |
| 28 |
266658_at |
AT2G25735
|
unknown |
0.409 |
0.065 |
| 29 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
0.398 |
0.459 |
| 30 |
266993_at |
AT2G39210
|
[Major facilitator superfamily protein] |
0.392 |
0.459 |
| 31 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.391 |
0.256 |
| 32 |
254253_at |
AT4G23320
|
CRK24, cysteine-rich RLK (RECEPTOR-like protein kinase) 24 |
0.391 |
0.244 |
| 33 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
0.388 |
0.705 |
| 34 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.383 |
0.356 |
| 35 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
0.382 |
0.136 |
| 36 |
253776_at |
AT4G28390
|
AAC3, ADP/ATP carrier 3, ATAAC3 |
0.377 |
0.185 |
| 37 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
0.356 |
0.355 |
| 38 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
0.350 |
0.160 |
| 39 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.339 |
0.571 |
| 40 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
0.330 |
0.148 |
| 41 |
251232_at |
AT3G62780
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.327 |
0.294 |
| 42 |
255627_at |
AT4G00955
|
unknown |
0.294 |
0.093 |
| 43 |
263457_at |
AT2G22300
|
CAMTA3, CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR 3, SR1, signal responsive 1 |
-0.654 |
0.086 |
| 44 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
-0.421 |
0.524 |
| 45 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.317 |
-0.126 |
| 46 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.185 |
-0.409 |
| 47 |
264053_at |
AT2G22560
|
NET2D, Networked 2D |
-0.182 |
0.136 |
| 48 |
250322_at |
AT5G12870
|
ATMYB46, MYB DOMAIN PROTEIN 46, MYB46, myb domain protein 46 |
-0.164 |
-0.156 |
| 49 |
259641_at |
AT1G69020
|
[Prolyl oligopeptidase family protein] |
-0.162 |
-0.071 |
| 50 |
256599_at |
AT3G14760
|
unknown |
-0.158 |
-0.371 |
| 51 |
261836_at |
AT1G16090
|
WAKL7, wall associated kinase-like 7 |
-0.156 |
0.067 |
| 52 |
246733_at |
AT5G27660
|
DEG14, degradation of periplasmic proteins 14 |
-0.149 |
-0.029 |
| 53 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
-0.146 |
0.352 |
| 54 |
250694_at |
AT5G06710
|
HAT14, homeobox from Arabidopsis thaliana |
-0.144 |
-0.221 |
| 55 |
265487_at |
AT2G15600
|
unknown |
-0.141 |
0.021 |
| 56 |
255420_at |
AT4G03240
|
ATFH, FH, frataxin homolog |
-0.139 |
0.028 |
| 57 |
247111_at |
AT5G65880
|
unknown |
-0.137 |
-0.103 |
| 58 |
247100_at |
AT5G66520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.137 |
-0.190 |
| 59 |
259235_at |
AT3G11600
|
unknown |
-0.137 |
0.083 |
| 60 |
246581_at |
AT1G31760
|
[SWIB/MDM2 domain superfamily protein] |
-0.136 |
-0.006 |
| 61 |
266278_at |
AT2G29300
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.135 |
-0.408 |
| 62 |
261261_at |
AT1G26730
|
[EXS (ERD1/XPR1/SYG1) family protein] |
-0.132 |
0.175 |
| 63 |
256302_at |
AT1G69526
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.130 |
-0.345 |
| 64 |
265865_at |
AT2G01740
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.124 |
-0.042 |
| 65 |
247022_at |
AT5G67050
|
[alpha/beta-Hydrolases superfamily protein] |
-0.123 |
0.027 |
| 66 |
266733_at |
AT2G03280
|
[O-fucosyltransferase family protein] |
-0.122 |
-0.015 |
| 67 |
244934_at |
ATCG01080
|
NDHG |
-0.122 |
-0.027 |
| 68 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.118 |
-0.232 |
| 69 |
259953_at |
AT1G74810
|
BOR5, REQUIRES HIGH BORON 5 |
-0.117 |
-0.033 |
| 70 |
257903_at |
AT3G28460
|
[methyltransferases] |
-0.117 |
-0.121 |
| 71 |
245078_at |
AT2G23340
|
DEAR3, DREB and EAR motif protein 3 |
-0.116 |
0.074 |
| 72 |
262972_at |
AT1G75620
|
[glyoxal oxidase-related protein] |
-0.116 |
-0.036 |
| 73 |
248653_at |
AT5G49290
|
ATRLP56, receptor like protein 56, RLP56, receptor like protein 56 |
-0.114 |
0.027 |
| 74 |
263040_at |
AT1G23300
|
[MATE efflux family protein] |
-0.114 |
-0.042 |
| 75 |
264602_at |
AT1G04700
|
[PB1 domain-containing protein tyrosine kinase] |
-0.113 |
0.034 |
| 76 |
262344_at |
AT1G64060
|
ATRBOH F, respiratory burst oxidase protein F, ATRBOHF, ARABIDOPSIS THALIANA RESPIRATORY BURST OXIDASE HOMOLOG F, RBOH F, respiratory burst oxidase protein F, RBOHAP108, RBOHF, RESPIRATORY BURST OXIDASE PROTEIN F |
-0.109 |
0.146 |
| 77 |
266736_at |
AT2G46960
|
CYP709B1, cytochrome P450, family 709, subfamily B, polypeptide 1 |
-0.108 |
-0.139 |
| 78 |
254288_at |
AT4G22970
|
AESP, homolog of separase, ESP, EXTRA SPINDLE POLES, ESP, homolog of separase, RSW4, RADIALLY SWOLLEN 4 |
-0.107 |
-0.049 |
| 79 |
248440_at |
AT5G51260
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.107 |
0.039 |
| 80 |
266357_at |
AT2G32290
|
BAM6, beta-amylase 6, BMY5, BETA-AMYLASE 5 |
-0.106 |
-0.253 |
| 81 |
259473_at |
AT1G19025
|
[DNA repair metallo-beta-lactamase family protein] |
-0.105 |
-0.069 |
| 82 |
255391_at |
AT4G03670
|
unknown |
-0.105 |
-0.020 |
| 83 |
260354_at |
AT1G69330
|
[RING/U-box superfamily protein] |
-0.104 |
-0.051 |