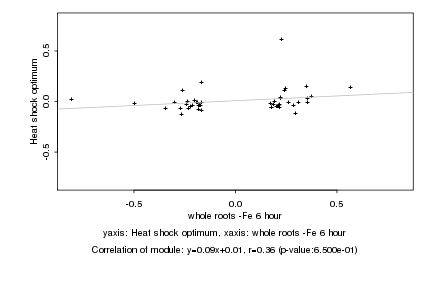
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
whole roots -Fe 6 hour |
Log2 signal ratio
Heat shock optimum |
| 1 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.566 |
0.139 |
| 2 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.372 |
0.056 |
| 3 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
0.351 |
0.000 |
| 4 |
253044_at |
AT4G37290
|
unknown |
0.350 |
0.035 |
| 5 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.347 |
0.156 |
| 6 |
264799_at |
AT1G08550
|
AVDE1, ARABIDOPSIS VIOLAXANTHIN DE-EPOXIDASE 1, NPQ1, non-photochemical quenching 1 |
0.309 |
-0.004 |
| 7 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
0.294 |
-0.109 |
| 8 |
245860_at |
AT5G28310
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.284 |
-0.038 |
| 9 |
264837_at |
AT1G03600
|
PSB27 |
0.261 |
-0.009 |
| 10 |
250670_at |
AT5G06860
|
ATPGIP1, POLYGALACTURONASE INHIBITING PROTEIN 1, PGIP1, polygalacturonase inhibiting protein 1 |
0.242 |
0.129 |
| 11 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.240 |
0.113 |
| 12 |
267459_at |
AT2G33850
|
unknown |
0.222 |
0.616 |
| 13 |
255870_at |
AT2G30280
|
DMS4, DEFECTIVE IN MERISTEM SILENCING 4, RDM4, RNA-directed DNA methylation 4 |
0.220 |
0.036 |
| 14 |
248299_at |
AT5G53080
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.219 |
0.040 |
| 15 |
249111_at |
AT5G43770
|
[proline-rich family protein] |
0.216 |
-0.027 |
| 16 |
261321_at |
AT1G44740
|
unknown |
0.213 |
-0.053 |
| 17 |
263965_at |
AT2G12870
|
[gypsy-like retrotransposon family, has a 3.8e-07 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
0.208 |
-0.035 |
| 18 |
260014_at |
AT1G68010
|
ATHPR1, HPR, hydroxypyruvate reductase |
0.205 |
-0.043 |
| 19 |
253742_at |
AT4G28900
|
[copia-like retrotransposon family, has a 7.7e-236 P-value blast match to GB:AAA57005 Hopscotch polyprotein (Ty1_Copia-element) (Zea mays)] |
0.200 |
-0.047 |
| 20 |
263020_at |
AT1G23880
|
[NHL domain-containing protein] |
0.192 |
0.005 |
| 21 |
257562_at |
AT3G30480
|
unknown |
0.185 |
-0.029 |
| 22 |
265796_at |
AT2G35730
|
[Heavy metal transport/detoxification superfamily protein ] |
0.174 |
-0.053 |
| 23 |
258748_at |
AT3G05930
|
GLP8, germin-like protein 8 |
0.174 |
-0.050 |
| 24 |
266516_at |
AT2G47880
|
[Glutaredoxin family protein] |
0.169 |
-0.010 |
| 25 |
257143_at |
AT3G20110
|
CYP705A20, cytochrome P450, family 705, subfamily A, polypeptide 20 |
-0.811 |
0.024 |
| 26 |
259454_at |
AT1G44050
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.501 |
-0.012 |
| 27 |
249364_at |
AT5G40590
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.349 |
-0.064 |
| 28 |
258541_at |
AT3G07000
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.303 |
-0.008 |
| 29 |
266765_at |
AT2G46860
|
AtPPa3, pyrophosphorylase 3, PPa3, pyrophosphorylase 3 |
-0.271 |
-0.065 |
| 30 |
247581_at |
AT5G61350
|
[Protein kinase superfamily protein] |
-0.269 |
-0.126 |
| 31 |
260987_at |
AT1G53590
|
NTMC2T6.1, NTMC2TYPE6.1 |
-0.264 |
0.111 |
| 32 |
249868_at |
AT5G23030
|
TET12, tetraspanin12 |
-0.244 |
-0.024 |
| 33 |
264698_at |
AT1G70200
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.241 |
0.004 |
| 34 |
264348_at |
AT1G12110
|
AtNPF6.3, ATNRT1, ARABIDOPSIS THALIANA NITRATE TRANSPORTER 1, B-1, CHL1, CHLORINA 1, CHL1-1, NPF6.3, NRT1/ PTR family 6.3, NRT1, NITRATE TRANSPORTER 1, NRT1.1, nitrate transporter 1.1 |
-0.232 |
-0.069 |
| 35 |
252205_at |
AT3G50350
|
unknown |
-0.226 |
-0.048 |
| 36 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
-0.215 |
-0.038 |
| 37 |
261318_at |
AT1G53035
|
unknown |
-0.206 |
0.011 |
| 38 |
260628_at |
AT1G62320
|
[ERD (early-responsive to dehydration stress) family protein] |
-0.194 |
-0.005 |
| 39 |
263536_at |
AT2G25000
|
ATWRKY60, WRKY60, WRKY DNA-binding protein 60 |
-0.187 |
-0.004 |
| 40 |
254545_at |
AT4G19830
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.184 |
-0.074 |
| 41 |
254575_at |
AT4G19460
|
[UDP-Glycosyltransferase superfamily protein] |
-0.183 |
-0.033 |
| 42 |
267276_at |
AT2G30130
|
ASL5, LBD12, PCK1, PEACOCK 1 |
-0.178 |
-0.035 |
| 43 |
261846_at |
AT1G11540
|
[Sulfite exporter TauE/SafE family protein] |
-0.178 |
-0.036 |
| 44 |
264029_at |
AT2G03720
|
MRH6, morphogenesis of root hair 6 |
-0.175 |
-0.033 |
| 45 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.172 |
0.196 |
| 46 |
254979_at |
AT4G10550
|
[Subtilase family protein] |
-0.172 |
-0.084 |
| 47 |
263763_at |
AT2G21385
|
unknown |
-0.169 |
-0.009 |