|
probeID |
AGICode |
Annotation |
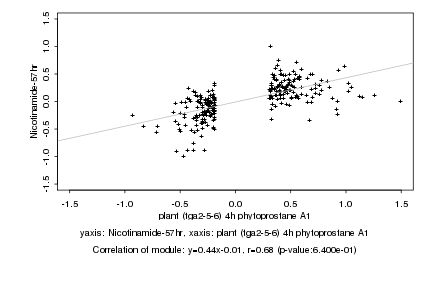
Log2 signal ratio
plant (tga2-5-6) 4h phytoprostane A1 |
Log2 signal ratio
Nicotinamide-57hr |
| 1 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
1.490 |
0.008 |
| 2 |
255811_at |
AT4G10250
|
ATHSP22.0 |
1.253 |
0.127 |
| 3 |
261838_at |
AT1G16030
|
Hsp70b, heat shock protein 70B |
1.143 |
0.090 |
| 4 |
253884_at |
AT4G27670
|
HSP21, heat shock protein 21 |
1.117 |
0.092 |
| 5 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
1.043 |
0.261 |
| 6 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
1.023 |
0.338 |
| 7 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
1.016 |
0.191 |
| 8 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
0.985 |
0.644 |
| 9 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.927 |
0.564 |
| 10 |
258695_at |
AT3G09640
|
APX1B, ASCORBATE PEROXIDASE 1B, APX2, ascorbate peroxidase 2 |
0.923 |
0.017 |
| 11 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
0.915 |
-0.228 |
| 12 |
266590_at |
AT2G46240
|
ATBAG6, ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 6, BAG6, BCL-2-associated athanogene 6 |
0.907 |
-0.135 |
| 13 |
262307_at |
AT1G71000
|
[Chaperone DnaJ-domain superfamily protein] |
0.874 |
0.056 |
| 14 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.849 |
0.262 |
| 15 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.832 |
0.375 |
| 16 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.776 |
0.382 |
| 17 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
0.773 |
0.201 |
| 18 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
0.757 |
0.302 |
| 19 |
265675_at |
AT2G32120
|
HSP70T-2, heat-shock protein 70T-2 |
0.756 |
0.139 |
| 20 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
0.724 |
0.161 |
| 21 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
0.721 |
0.245 |
| 22 |
246018_at |
AT5G10695
|
unknown |
0.716 |
0.323 |
| 23 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.695 |
0.498 |
| 24 |
263164_at |
AT1G03070
|
AtLFG4, LFG4, LIFEGUARD 4 |
0.692 |
0.083 |
| 25 |
254062_at |
AT4G25380
|
AtSAP10, Arabidopsis thaliana stress-associated protein 10, SAP10, stress-associated protein 10 |
0.685 |
0.234 |
| 26 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
0.683 |
-0.004 |
| 27 |
260406_at |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
0.677 |
0.495 |
| 28 |
257421_at |
AT1G12030
|
unknown |
0.664 |
-0.338 |
| 29 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.649 |
-0.013 |
| 30 |
249770_at |
AT5G24110
|
ATWRKY30, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 30, WRKY30, WRKY DNA-binding protein 30 |
0.643 |
0.430 |
| 31 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
0.639 |
0.117 |
| 32 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.595 |
0.585 |
| 33 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
0.595 |
0.140 |
| 34 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.590 |
0.269 |
| 35 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.576 |
0.457 |
| 36 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.572 |
0.446 |
| 37 |
261564_at |
AT1G01720
|
ANAC002, Arabidopsis NAC domain containing protein 2, ATAF1 |
0.571 |
0.414 |
| 38 |
259831_at |
AT1G69600
|
ATHB29, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 29, ZFHD1, zinc finger homeodomain 1, ZHD11, ZINC FINGER HOMEODOMAIN 11 |
0.570 |
0.072 |
| 39 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.559 |
0.422 |
| 40 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
0.557 |
0.105 |
| 41 |
260803_at |
AT1G78340
|
ATGSTU22, glutathione S-transferase TAU 22, GSTU22, glutathione S-transferase TAU 22 |
0.551 |
0.112 |
| 42 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
0.545 |
0.714 |
| 43 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
0.542 |
0.493 |
| 44 |
267263_at |
AT2G23110
|
[Late embryogenesis abundant protein, group 6] |
0.542 |
0.084 |
| 45 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.533 |
0.411 |
| 46 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
0.532 |
0.260 |
| 47 |
259037_at |
AT3G09350
|
Fes1A, Fes1A |
0.524 |
0.173 |
| 48 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
0.524 |
0.367 |
| 49 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
0.521 |
0.552 |
| 50 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
0.508 |
0.285 |
| 51 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
0.502 |
0.078 |
| 52 |
261956_at |
AT1G64590
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.501 |
0.062 |
| 53 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.499 |
0.293 |
| 54 |
255543_at |
AT4G01870
|
[tolB protein-related] |
0.497 |
0.203 |
| 55 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.489 |
0.499 |
| 56 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
0.489 |
0.350 |
| 57 |
258939_at |
AT3G10020
|
unknown |
0.488 |
-0.072 |
| 58 |
251884_at |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.487 |
0.392 |
| 59 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.484 |
0.214 |
| 60 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
0.476 |
0.403 |
| 61 |
264746_at |
AT1G62300
|
ATWRKY6, WRKY6 |
0.474 |
0.318 |
| 62 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
0.471 |
0.302 |
| 63 |
257226_at |
AT3G27880
|
unknown |
0.463 |
0.157 |
| 64 |
266296_at |
AT2G29420
|
ATGSTU7, glutathione S-transferase tau 7, GST25, GLUTATHIONE S-TRANSFERASE 25, GSTU7, glutathione S-transferase tau 7 |
0.462 |
0.233 |
| 65 |
264929_at |
AT1G60730
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.459 |
0.266 |
| 66 |
257216_at |
AT3G14990
|
AtDJ1A, DJ-1 homolog A, DJ-1a, DJ1A, DJ-1 homolog A |
0.456 |
0.215 |
| 67 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
0.455 |
-0.042 |
| 68 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.454 |
0.288 |
| 69 |
245243_at |
AT1G44414
|
unknown |
0.452 |
0.203 |
| 70 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.447 |
0.206 |
| 71 |
263073_at |
AT2G17500
|
[Auxin efflux carrier family protein] |
0.444 |
0.188 |
| 72 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.444 |
0.481 |
| 73 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.442 |
0.239 |
| 74 |
260261_at |
AT1G68450
|
PDE337, PIGMENT DEFECTIVE 337 |
0.442 |
0.395 |
| 75 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.439 |
0.197 |
| 76 |
264580_at |
AT1G05340
|
unknown |
0.433 |
0.491 |
| 77 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
0.432 |
0.137 |
| 78 |
254263_at |
AT4G23493
|
unknown |
0.432 |
0.067 |
| 79 |
258203_at |
AT3G13950
|
unknown |
0.430 |
0.491 |
| 80 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.424 |
0.269 |
| 81 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.421 |
0.279 |
| 82 |
259979_at |
AT1G76600
|
unknown |
0.417 |
0.268 |
| 83 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
0.416 |
0.505 |
| 84 |
247691_at |
AT5G59720
|
HSP18.2, heat shock protein 18.2 |
0.409 |
-0.025 |
| 85 |
246988_at |
AT5G67340
|
[ARM repeat superfamily protein] |
0.408 |
0.353 |
| 86 |
252045_at |
AT3G52450
|
AtPUB22, PUB22, plant U-box 22 |
0.407 |
0.502 |
| 87 |
261265_at |
AT1G26800
|
[RING/U-box superfamily protein] |
0.406 |
0.073 |
| 88 |
245560_at |
AT4G15480
|
UGT84A1 |
0.405 |
0.138 |
| 89 |
262656_at |
AT1G14200
|
[RING/U-box superfamily protein] |
0.403 |
0.191 |
| 90 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.401 |
0.567 |
| 91 |
256324_at |
AT1G66760
|
[MATE efflux family protein] |
0.399 |
0.112 |
| 92 |
254759_at |
AT4G13180
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.398 |
0.297 |
| 93 |
246944_at |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
0.397 |
0.322 |
| 94 |
247351_at |
AT5G63790
|
ANAC102, NAC domain containing protein 102, NAC102, NAC domain containing protein 102 |
0.395 |
0.181 |
| 95 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.391 |
0.204 |
| 96 |
259120_at |
AT3G02240
|
GLV4, GOLVEN 4, RGF7, root meristem growth factor 7 |
0.386 |
0.761 |
| 97 |
265660_at |
AT2G25470
|
AtRLP21, receptor like protein 21, RLP21, receptor like protein 21 |
0.380 |
0.040 |
| 98 |
263475_at |
AT2G31945
|
unknown |
0.379 |
0.656 |
| 99 |
250252_at |
AT5G13750
|
ZIFL1, zinc induced facilitator-like 1 |
0.379 |
0.268 |
| 100 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.378 |
0.276 |
| 101 |
246612_at |
AT5G35320
|
unknown |
0.368 |
0.195 |
| 102 |
253830_at |
AT4G27652
|
unknown |
0.368 |
0.389 |
| 103 |
261748_at |
AT1G76070
|
unknown |
0.365 |
0.417 |
| 104 |
245313_at |
AT4G15420
|
[Ubiquitin fusion degradation UFD1 family protein] |
0.364 |
0.126 |
| 105 |
266514_at |
AT2G47890
|
[B-box type zinc finger protein with CCT domain] |
0.362 |
-0.087 |
| 106 |
245272_at |
AT4G17250
|
unknown |
0.359 |
0.117 |
| 107 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.358 |
0.605 |
| 108 |
246325_at |
AT1G16540
|
ABA3, ABA DEFICIENT 3, ACI2, ALTERED CHLOROPLAST IMPORT 2, ATABA3, AtLOS5, LOS5, LOW OSMOTIC STRESS 5, SIR3, SIRTINOL RESISTANT 3 |
0.354 |
0.117 |
| 109 |
259479_at |
AT1G19020
|
unknown |
0.352 |
0.413 |
| 110 |
248934_at |
AT5G46080
|
[Protein kinase superfamily protein] |
0.351 |
0.483 |
| 111 |
251752_at |
AT3G55740
|
ATPROT2, PROLINE TRANSPORTER 2, PROT2, proline transporter 2 |
0.348 |
0.235 |
| 112 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
0.348 |
0.246 |
| 113 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
0.344 |
0.443 |
| 114 |
263433_at |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
0.333 |
-0.052 |
| 115 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
0.333 |
0.059 |
| 116 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.333 |
0.507 |
| 117 |
260602_at |
AT1G55920
|
ATSERAT2;1, serine acetyltransferase 2;1, SAT1, SERINE ACETYLTRANSFERASE 1, SAT5, SERINE ACETYLTRANSFERASE 5, SERAT2;1, serine acetyltransferase 2;1 |
0.329 |
-0.038 |
| 118 |
254432_at |
AT4G20830
|
[FAD-binding Berberine family protein] |
0.327 |
0.247 |
| 119 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.325 |
0.338 |
| 120 |
260039_at |
AT1G68795
|
CLE12, CLAVATA3/ESR-RELATED 12 |
0.325 |
-0.142 |
| 121 |
256285_at |
AT3G12510
|
[MADS-box family protein] |
0.324 |
0.295 |
| 122 |
247509_at |
AT5G62020
|
AT-HSFB2A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR B2A, HSFB2A, heat shock transcription factor B2A |
0.322 |
0.226 |
| 123 |
248381_at |
AT5G51830
|
[pfkB-like carbohydrate kinase family protein] |
0.322 |
0.296 |
| 124 |
251130_at |
AT5G01180
|
AtNPF8.2, ATPTR5, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 5, NPF8.2, NRT1/ PTR family 8.2, PTR5, peptide transporter 5 |
0.320 |
-0.312 |
| 125 |
249377_at |
AT5G40690
|
unknown |
0.320 |
0.246 |
| 126 |
245685_at |
AT5G22220
|
ATE2FB, E2F1, E2F transcription factor 1, E2FB |
0.320 |
0.099 |
| 127 |
267372_at |
AT2G26290
|
ARSK1, root-specific kinase 1 |
0.314 |
0.199 |
| 128 |
249721_at |
AT5G24655
|
LSU4, RESPONSE TO LOW SULFUR 4 |
0.314 |
0.060 |
| 129 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.313 |
1.011 |
| 130 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.312 |
0.120 |
| 131 |
247488_at |
AT5G61820
|
unknown |
0.311 |
0.215 |
| 132 |
250493_at |
AT5G09800
|
[ARM repeat superfamily protein] |
0.308 |
0.219 |
| 133 |
265008_at |
AT1G61560
|
ATMLO6, MILDEW RESISTANCE LOCUS O 6, MLO6, MILDEW RESISTANCE LOCUS O 6 |
0.308 |
0.068 |
| 134 |
245126_at |
AT2G47460
|
ATMYB12, MYB DOMAIN PROTEIN 12, MYB12, myb domain protein 12, PFG1, PRODUCTION OF FLAVONOL GLYCOSIDES 1 |
0.308 |
0.066 |
| 135 |
246229_at |
AT4G37160
|
sks15, SKU5 similar 15 |
-0.939 |
-0.237 |
| 136 |
247914_at |
AT5G57540
|
AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 |
-0.838 |
-0.437 |
| 137 |
256352_at |
AT1G54970
|
ATPRP1, proline-rich protein 1, PRP1, proline-rich protein 1, RHS7, ROOT HAIR SPECIFIC 7 |
-0.721 |
-0.554 |
| 138 |
267287_at |
AT2G23630
|
sks16, SKU5 similar 16 |
-0.710 |
-0.447 |
| 139 |
263376_at |
AT2G20520
|
FLA6, FASCICLIN-like arabinogalactan 6 |
-0.569 |
-0.194 |
| 140 |
264775_at |
AT1G22880
|
ATCEL5, ARABIDOPSIS THALIANA CELLULASE 5, ATGH9B4, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B4, CEL5, cellulase 5 |
-0.547 |
-0.019 |
| 141 |
262105_at |
AT1G02810
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.545 |
-0.361 |
| 142 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.538 |
-0.903 |
| 143 |
255695_at |
AT4G00080
|
UNE11, unfertilized embryo sac 11 |
-0.521 |
-0.414 |
| 144 |
253763_at |
AT4G28850
|
ATXTH26, XTH26, xyloglucan endotransglucosylase/hydrolase 26 |
-0.512 |
-0.494 |
| 145 |
250469_at |
AT5G10130
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.499 |
-0.527 |
| 146 |
263232_at |
AT1G05700
|
[Leucine-rich repeat transmembrane protein kinase protein] |
-0.498 |
-0.210 |
| 147 |
260558_at |
AT2G43600
|
[Chitinase family protein] |
-0.491 |
-0.016 |
| 148 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.475 |
-0.985 |
| 149 |
253483_at |
AT4G31910
|
BAT1, BR-related AcylTransferase1, PIZ, PIZZA |
-0.465 |
-0.225 |
| 150 |
247634_at |
AT5G60520
|
[Late embryogenesis abundant (LEA) protein-related] |
-0.464 |
-0.289 |
| 151 |
249203_at |
AT5G42590
|
CYP71A16, cytochrome P450, family 71, subfamily A, polypeptide 16, MRO, marneral oxidase |
-0.457 |
-0.004 |
| 152 |
252537_at |
AT3G45710
|
[Major facilitator superfamily protein] |
-0.452 |
-0.423 |
| 153 |
263970_at |
AT2G42850
|
CYP718, cytochrome P450, family 718 |
-0.450 |
-0.094 |
| 154 |
253936_at |
AT4G26880
|
[Stigma-specific Stig1 family protein] |
-0.440 |
0.061 |
| 155 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.436 |
-0.882 |
| 156 |
253618_at |
AT4G30420
|
UMAMIT34, Usually multiple acids move in and out Transporters 34 |
-0.427 |
0.238 |
| 157 |
252988_at |
AT4G38410
|
[Dehydrin family protein] |
-0.423 |
0.044 |
| 158 |
259264_at |
AT3G01260
|
[Galactose mutarotase-like superfamily protein] |
-0.408 |
0.001 |
| 159 |
247094_at |
AT5G66280
|
GMD1, GDP-D-mannose 4,6-dehydratase 1 |
-0.398 |
-0.514 |
| 160 |
261562_at |
AT1G01750
|
ADF11, actin depolymerizing factor 11 |
-0.388 |
-0.884 |
| 161 |
247074_at |
AT5G66590
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.387 |
-0.306 |
| 162 |
259596_at |
AT1G28130
|
GH3.17 |
-0.382 |
0.194 |
| 163 |
260758_at |
AT1G48930
|
AtGH9C1, glycosyl hydrolase 9C1, GH9C1, glycosyl hydrolase 9C1 |
-0.380 |
-0.745 |
| 164 |
261099_at |
AT1G62980
|
ATEXP18, ATEXPA18, expansin A18, ATHEXP ALPHA 1.25, EXP18, EXPANSIN 18, EXPA18, expansin A18 |
-0.379 |
-0.547 |
| 165 |
256774_at |
AT3G13760
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.369 |
0.126 |
| 166 |
266765_at |
AT2G46860
|
AtPPa3, pyrophosphorylase 3, PPa3, pyrophosphorylase 3 |
-0.367 |
-0.285 |
| 167 |
256349_at |
AT1G54890
|
[Late embryogenesis abundant (LEA) protein-related] |
-0.366 |
0.165 |
| 168 |
253353_at |
AT4G33730
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.360 |
-0.423 |
| 169 |
262666_at |
AT1G14080
|
ATFUT6, FUT6, fucosyltransferase 6 |
-0.357 |
-0.246 |
| 170 |
245966_at |
AT5G19790
|
RAP2.11, related to AP2 11 |
-0.353 |
-0.343 |
| 171 |
259854_at |
AT1G72200
|
[RING/U-box superfamily protein] |
-0.352 |
-0.514 |
| 172 |
248408_at |
AT5G51520
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.351 |
0.103 |
| 173 |
247922_at |
AT5G57500
|
[Galactosyltransferase family protein] |
-0.347 |
-0.108 |
| 174 |
259735_at |
AT1G64405
|
unknown |
-0.345 |
0.002 |
| 175 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
-0.345 |
-0.273 |
| 176 |
256360_at |
AT1G66440
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.335 |
0.006 |
| 177 |
265586_at |
AT2G19990
|
PR-1-LIKE, pathogenesis-related protein-1-like |
-0.327 |
-0.237 |
| 178 |
247476_at |
AT5G62330
|
unknown |
-0.325 |
0.113 |
| 179 |
251434_at |
AT3G59850
|
[Pectin lyase-like superfamily protein] |
-0.318 |
-0.023 |
| 180 |
245555_at |
AT4G15390
|
[HXXXD-type acyl-transferase family protein] |
-0.317 |
-0.013 |
| 181 |
246807_at |
AT5G27100
|
ATGLR2.1, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 2.1, GLR2.1, glutamate receptor 2.1 |
-0.317 |
0.053 |
| 182 |
265102_at |
AT1G30870
|
[Peroxidase superfamily protein] |
-0.315 |
-0.621 |
| 183 |
247097_at |
AT5G66460
|
AtMAN7, MAN7, endo-beta-mannase 7 |
-0.314 |
-0.383 |
| 184 |
252510_at |
AT3G46270
|
[receptor protein kinase-related] |
-0.310 |
0.089 |
| 185 |
261550_at |
AT1G63450
|
RHS8, root hair specific 8, XUT1, XYLOGLUCAN-SPECIFIC GALACTURONOSYLTRANSFERASE 1 |
-0.305 |
-0.096 |
| 186 |
263373_at |
AT2G20515
|
unknown |
-0.304 |
-0.392 |
| 187 |
266920_at |
AT2G45750
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.304 |
-0.310 |
| 188 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
-0.304 |
-0.221 |
| 189 |
253227_at |
AT4G35030
|
[Protein kinase superfamily protein] |
-0.303 |
-0.183 |
| 190 |
260230_at |
AT1G74500
|
ATBS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, bHLH135, basic helix-loop-helix protein 135, BS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, PRE3, PACLOBUTRAZOL RESISTANCE 3, TMO7, TARGET OF MONOPTEROS 7 |
-0.302 |
-0.474 |
| 191 |
252172_at |
AT3G50640
|
unknown |
-0.302 |
-0.354 |
| 192 |
264804_at |
AT1G08590
|
[Leucine-rich receptor-like protein kinase family protein] |
-0.300 |
-0.056 |
| 193 |
250224_at |
AT5G14150
|
unknown |
-0.300 |
-0.365 |
| 194 |
248652_at |
AT5G49270
|
COBL9, COBRA-LIKE 9, DER9, DEFORMED ROOT HAIRS 9, MRH4, MUTANT ROOT HAIR 4, SHV2, SHAVEN 2 |
-0.296 |
-0.253 |
| 195 |
251255_at |
AT3G62280
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.289 |
-0.316 |
| 196 |
259813_at |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
-0.284 |
-0.878 |
| 197 |
262915_at |
AT1G59940
|
ARR3, response regulator 3 |
-0.284 |
-0.176 |
| 198 |
257467_at |
AT1G31320
|
LBD4, LOB domain-containing protein 4 |
-0.280 |
-0.226 |
| 199 |
248406_at |
AT5G51490
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.276 |
0.041 |
| 200 |
251116_at |
AT3G63470
|
scpl40, serine carboxypeptidase-like 40 |
-0.275 |
-0.002 |
| 201 |
247352_at |
AT5G63650
|
SNRK2-5, SNRK2.5, SNF1-related protein kinase 2.5, SRK2H, SNF1-RELATED PROTEIN KINASE 2H |
-0.274 |
-0.376 |
| 202 |
251293_at |
AT3G61930
|
unknown |
-0.273 |
0.035 |
| 203 |
258421_at |
AT3G16690
|
AtSWEET16, SWEET16 |
-0.272 |
-0.187 |
| 204 |
267307_at |
AT2G30210
|
LAC3, laccase 3 |
-0.272 |
0.037 |
| 205 |
246390_at |
AT1G77330
|
ACO5, ACC oxidase 5 |
-0.265 |
-0.197 |
| 206 |
246540_at |
AT5G15600
|
SP1L4, SPIRAL1-like4 |
-0.264 |
-0.033 |
| 207 |
253786_at |
AT4G28650
|
[Leucine-rich repeat transmembrane protein kinase family protein] |
-0.263 |
-0.242 |
| 208 |
246562_at |
AT5G15580
|
LNG1, LONGIFOLIA1, TRM2, TON1 Recruiting Motif 2 |
-0.262 |
-0.183 |
| 209 |
247474_at |
AT5G62280
|
unknown |
-0.262 |
-0.054 |
| 210 |
267410_at |
AT2G34920
|
EDA18, embryo sac development arrest 18 |
-0.259 |
-0.158 |
| 211 |
245586_at |
AT4G14980
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.257 |
0.027 |
| 212 |
259525_at |
AT1G12560
|
ATEXP7, ATEXPA7, expansin A7, ATHEXP ALPHA 1.26, EXP7, EXPA7, expansin A7 |
-0.257 |
-0.433 |
| 213 |
258700_at |
AT3G09710
|
IQD1, IQ-domain 1 |
-0.251 |
-0.059 |
| 214 |
252568_at |
AT3G45410
|
LecRK-I.3, L-type lectin receptor kinase I.3 |
-0.250 |
0.190 |
| 215 |
257197_at |
AT3G23800
|
SBP3, selenium-binding protein 3 |
-0.249 |
-0.021 |
| 216 |
263809_at |
AT2G04570
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.248 |
0.069 |
| 217 |
260539_at |
AT2G43480
|
[Peroxidase superfamily protein] |
-0.248 |
-0.005 |
| 218 |
256926_at |
AT3G22540
|
unknown |
-0.247 |
-0.172 |
| 219 |
249773_at |
AT5G24140
|
SQP2, squalene monooxygenase 2 |
-0.246 |
-0.060 |
| 220 |
257613_at |
AT3G26610
|
[Pectin lyase-like superfamily protein] |
-0.245 |
-0.112 |
| 221 |
249055_at |
AT5G44460
|
CML43, calmodulin like 43 |
-0.244 |
0.030 |
| 222 |
263146_at |
AT1G53940
|
GLIP2, GDSL-motif lipase 2 |
-0.241 |
-0.030 |
| 223 |
263596_at |
AT2G01900
|
[DNAse I-like superfamily protein] |
-0.239 |
-0.073 |
| 224 |
246913_at |
AT5G25830
|
GATA12, GATA transcription factor 12 |
-0.238 |
-0.197 |
| 225 |
256137_at |
AT1G48690
|
[Auxin-responsive GH3 family protein] |
-0.238 |
-0.001 |
| 226 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.231 |
-0.266 |
| 227 |
249227_at |
AT5G42180
|
PER64, peroxidase 64 |
-0.229 |
-0.251 |
| 228 |
254522_at |
AT4G19980
|
unknown |
-0.228 |
-0.086 |
| 229 |
247284_at |
AT5G64410
|
ATOPT4, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 4, OPT4, oligopeptide transporter 4 |
-0.227 |
0.118 |
| 230 |
260553_at |
AT2G41800
|
unknown |
-0.226 |
0.116 |
| 231 |
252964_at |
AT4G38830
|
CRK26, cysteine-rich RLK (RECEPTOR-like protein kinase) 26 |
-0.226 |
0.011 |
| 232 |
247179_at |
AT5G65320
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.225 |
-0.002 |
| 233 |
261999_at |
AT1G33800
|
AtGXMT1, GXM1, glucuronoxylan methyltransferase 1, GXMT1, glucuronoxylan methyltransferase 1 |
-0.224 |
-0.221 |
| 234 |
255528_at |
AT4G02090
|
unknown |
-0.218 |
-0.023 |
| 235 |
259897_at |
AT1G71380
|
ATCEL3, cellulase 3, ATGH9B3, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B3, CEL3, cellulase 3 |
-0.217 |
-0.029 |
| 236 |
252224_at |
AT3G49860
|
ARLA1B, ADP-ribosylation factor-like A1B, ATARLA1B, ADP-ribosylation factor-like A1B |
-0.216 |
-0.097 |
| 237 |
264941_at |
AT1G60680
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.214 |
0.214 |
| 238 |
249522_at |
AT5G38700
|
unknown |
-0.210 |
0.083 |
| 239 |
248003_at |
AT5G56220
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.208 |
-0.153 |
| 240 |
263151_at |
AT1G54120
|
unknown |
-0.208 |
-0.116 |
| 241 |
258719_at |
AT3G09540
|
[Pectin lyase-like superfamily protein] |
-0.207 |
-0.479 |
| 242 |
249167_at |
AT5G42860
|
unknown |
-0.203 |
-0.468 |
| 243 |
254392_at |
AT4G21600
|
ENDO5, endonuclease 5 |
-0.201 |
-0.089 |
| 244 |
265099_at |
AT1G03990
|
[Long-chain fatty alcohol dehydrogenase family protein] |
-0.201 |
0.139 |
| 245 |
267618_at |
AT2G26760
|
CYCB1;4, Cyclin B1;4 |
-0.200 |
-0.328 |
| 246 |
248681_at |
AT5G48900
|
[Pectin lyase-like superfamily protein] |
-0.200 |
-0.207 |
| 247 |
261653_at |
AT1G01900
|
ATSBT1.1, SBTI1.1 |
-0.199 |
-0.125 |
| 248 |
258905_at |
AT3G06390
|
[Uncharacterised protein family (UPF0497)] |
-0.199 |
0.090 |
| 249 |
261350_at |
AT1G79770
|
unknown |
-0.197 |
0.292 |
| 250 |
254193_at |
AT4G23870
|
unknown |
-0.197 |
-0.159 |
| 251 |
248656_at |
AT5G48460
|
[Actin binding Calponin homology (CH) domain-containing protein] |
-0.196 |
-0.497 |
| 252 |
257204_at |
AT3G23805
|
RALFL24, ralf-like 24 |
-0.196 |
-0.126 |
| 253 |
248570_at |
AT5G49780
|
[Leucine-rich repeat protein kinase family protein] |
-0.195 |
-0.009 |
| 254 |
253054_at |
AT4G37580
|
COP3, CONSTITUTIVE PHOTOMORPHOGENIC 3, HLS1, HOOKLESS 1, UNS2, UNUSUAL SUGAR RESPONSE 2 |
-0.195 |
0.060 |
| 255 |
250277_at |
AT5G12940
|
[Leucine-rich repeat (LRR) family protein] |
-0.194 |
-0.156 |
| 256 |
249768_at |
AT5G24100
|
[Leucine-rich repeat protein kinase family protein] |
-0.194 |
0.019 |
| 257 |
255345_at |
AT4G04460
|
[Saposin-like aspartyl protease family protein] |
-0.194 |
0.075 |
| 258 |
261924_at |
AT1G22550
|
[Major facilitator superfamily protein] |
-0.194 |
-0.056 |
| 259 |
258540_at |
AT3G06990
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.193 |
0.032 |
| 260 |
256673_at |
AT3G52370
|
FLA15, FASCICLIN-like arabinogalactan protein 15 precursor |
-0.193 |
-0.314 |
| 261 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
-0.192 |
-0.283 |
| 262 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
-0.192 |
0.341 |
| 263 |
253811_at |
AT4G28190
|
ULT, ULTRAPETALA, ULT1, ULTRAPETALA1 |
-0.192 |
-0.065 |
| 264 |
248822_at |
AT5G47000
|
[Peroxidase superfamily protein] |
-0.191 |
-0.200 |
| 265 |
253255_at |
AT4G34760
|
SAUR50, SMALL AUXIN UPREGULATED RNA 50 |
-0.190 |
0.046 |
| 266 |
262819_at |
AT1G11600
|
CYP77B1, cytochrome P450, family 77, subfamily B, polypeptide 1 |
-0.190 |
-0.175 |
| 267 |
251231_at |
AT3G62760
|
ATGSTF13 |
-0.189 |
-0.061 |