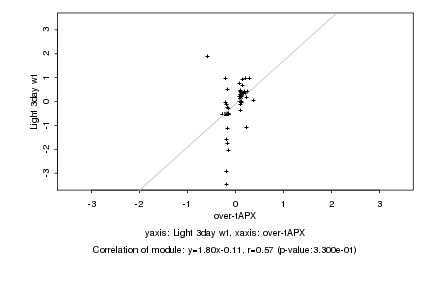
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
over-tAPX |
Log2 signal ratio
Light 3day wt |
| 1 |
259707_at |
AT1G77490
|
TAPX, thylakoidal ascorbate peroxidase |
0.366 |
0.046 |
| 2 |
250696_at |
AT5G06790
|
unknown |
0.288 |
0.981 |
| 3 |
245364_at |
AT4G15790
|
unknown |
0.242 |
0.421 |
| 4 |
259850_at |
AT1G72240
|
unknown |
0.218 |
-1.062 |
| 5 |
250844_at |
AT5G04470
|
SIM, SIAMESE |
0.215 |
0.194 |
| 6 |
257081_at |
AT3G30460
|
[RING/U-box superfamily protein] |
0.202 |
1.000 |
| 7 |
251490_at |
AT3G59490
|
unknown |
0.189 |
0.356 |
| 8 |
256723_at |
AT2G34160
|
[Alba DNA/RNA-binding protein] |
0.181 |
0.429 |
| 9 |
261421_at |
AT1G18840
|
IQD30, IQ-domain 30 |
0.168 |
0.383 |
| 10 |
254130_at |
AT4G24540
|
AGL24, AGAMOUS-like 24 |
0.165 |
0.346 |
| 11 |
246144_at |
AT5G20110
|
[Dynein light chain type 1 family protein] |
0.164 |
0.376 |
| 12 |
262533_at |
AT1G17090
|
unknown |
0.139 |
0.708 |
| 13 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
0.133 |
0.938 |
| 14 |
256052_at |
AT1G06960
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.125 |
0.387 |
| 15 |
259889_at |
AT1G76405
|
unknown |
0.125 |
0.354 |
| 16 |
245665_at |
AT1G28250
|
unknown |
0.119 |
0.269 |
| 17 |
255420_at |
AT4G03240
|
ATFH, FH, frataxin homolog |
0.117 |
0.404 |
| 18 |
253474_at |
AT4G32270
|
[Ubiquitin-like superfamily protein] |
0.116 |
0.387 |
| 19 |
252388_at |
AT3G47850
|
unknown |
0.113 |
-0.007 |
| 20 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.111 |
0.030 |
| 21 |
260650_at |
AT1G32370
|
TOM2B, tobamovirus multiplication 2B, TTM1 |
0.109 |
0.250 |
| 22 |
260129_at |
AT1G36380
|
unknown |
0.102 |
0.493 |
| 23 |
262434_at |
AT1G47670
|
[Transmembrane amino acid transporter family protein] |
0.102 |
-0.362 |
| 24 |
254487_at |
AT4G20780
|
CML42, calmodulin like 42 |
0.100 |
0.164 |
| 25 |
264098_at |
AT1G79260
|
unknown |
0.099 |
0.154 |
| 26 |
252426_at |
AT3G47630
|
unknown |
0.097 |
0.323 |
| 27 |
264550_at |
AT1G09450
|
AtHaspin, Haspin, Haspin-related gene |
0.097 |
0.322 |
| 28 |
267462_at |
AT2G33735
|
[Chaperone DnaJ-domain superfamily protein] |
0.096 |
0.422 |
| 29 |
254226_at |
AT4G23690
|
AtDIR6, Arabidopsis thaliana dirigent protein 6, DIR6, dirigent protein 6 |
0.094 |
-0.110 |
| 30 |
265142_at |
AT1G51360
|
ATDABB1, DIMERIC A/B BARREL DOMAINS-PROTEIN 1, DABB1, dimeric A/B barrel domainS-protein 1 |
0.092 |
0.135 |
| 31 |
250072_at |
AT5G17210
|
unknown |
0.091 |
0.037 |
| 32 |
251986_at |
AT3G53310
|
[AP2/B3-like transcriptional factor family protein] |
0.082 |
0.037 |
| 33 |
253219_at |
AT4G34990
|
AtMYB32, myb domain protein 32, MYB32, myb domain protein 32 |
0.080 |
0.794 |
| 34 |
245097_at |
AT2G40935
|
[PLAC8 family protein] |
0.079 |
0.221 |
| 35 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
-0.585 |
1.906 |
| 36 |
265949_at |
AT2G18540
|
[RmlC-like cupins superfamily protein] |
-0.289 |
-0.515 |
| 37 |
257346_at |
AT3G30846
|
[gypsy-like retrotransposon family, has a 1.0e-303 P-value blast match to GB:AAD27547 polyprotein (Gypsy_Ty3-element) (Oryza sativa subsp. indica)] |
-0.275 |
-0.515 |
| 38 |
255534_at |
AT4G02190
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.244 |
-0.515 |
| 39 |
261186_at |
AT1G34530
|
[CACTA-like transposase family (Ptta/En/Spm), has a 7.8e-116 P-value blast match to At5g36655.1/81-333 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.232 |
-0.515 |
| 40 |
255399_at |
AT4G03750
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.224 |
-0.515 |
| 41 |
260921_at |
AT1G21540
|
[AMP-dependent synthetase and ligase family protein] |
-0.215 |
-0.022 |
| 42 |
245804_at |
AT1G46696
|
unknown |
-0.213 |
-0.515 |
| 43 |
252843_at |
AT3G42130
|
[glycine-rich protein] |
-0.213 |
-0.515 |
| 44 |
256584_at |
AT3G28750
|
unknown |
-0.209 |
0.978 |
| 45 |
265211_at |
AT2G36640
|
ATECP63, embryonic cell protein 63, ECP63, embryonic cell protein 63 |
-0.208 |
-0.515 |
| 46 |
257950_at |
AT3G21780
|
UGT71B6, UDP-glucosyl transferase 71B6 |
-0.203 |
-1.584 |
| 47 |
245945_at |
AT5G19560
|
ATROPGEF10, ARABIDOPSIS THALIANA ROP UANINE NUCLEOTIDE EXCHANGE FACTOR 10, GEF10, guanine exchange factor 10, ROPGEF10, ROP (rho of plants) guanine nucleotide exchange factor 10 |
-0.195 |
-0.538 |
| 48 |
256926_at |
AT3G22540
|
unknown |
-0.194 |
-0.113 |
| 49 |
257591_at |
AT3G24900
|
AtRLP39, receptor like protein 39, RLP39, receptor like protein 39 |
-0.189 |
-3.432 |
| 50 |
258942_at |
AT3G09960
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.188 |
-2.902 |
| 51 |
248470_at |
AT5G50830
|
unknown |
-0.188 |
-0.515 |
| 52 |
245717_at |
AT5G33390
|
[glycine-rich protein] |
-0.182 |
-0.515 |
| 53 |
266493_at |
AT2G07030
|
[Mutator-like transposase family, has a 2.9e-11 P-value blast match to Q9XE24 /118-277 Pfam PF03108 MuDR family transposase (MuDr-element domain)] |
-0.181 |
-0.515 |
| 54 |
267193_at |
AT2G30900
|
TBL43, TRICHOME BIREFRINGENCE-LIKE 43 |
-0.177 |
-0.242 |
| 55 |
245076_at |
AT2G23170
|
GH3.3 |
-0.176 |
-1.103 |
| 56 |
259956_at |
AT1G75110
|
RRA2, REDUCED RESIDUAL ARABINOSE 2 |
-0.176 |
0.541 |
| 57 |
259027_at |
AT3G09280
|
unknown |
-0.171 |
-1.752 |
| 58 |
264332_at |
AT1G61920
|
unknown |
-0.169 |
-0.515 |
| 59 |
259747_at |
AT1G71170
|
[6-phosphogluconate dehydrogenase family protein] |
-0.164 |
-0.476 |
| 60 |
260890_at |
AT1G29090
|
[Cysteine proteinases superfamily protein] |
-0.162 |
-0.515 |
| 61 |
254841_at |
AT4G11940
|
unknown |
-0.162 |
-0.515 |
| 62 |
256197_at |
AT1G36910
|
[CACTA-like transposase family (Ptta/En/Spm), has a 4.0e-81 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.159 |
-0.515 |
| 63 |
246782_at |
AT5G27320
|
ATGID1C, GID1C, GA INSENSITIVE DWARF1C |
-0.158 |
-0.269 |
| 64 |
252627_at |
AT3G44950
|
[glycine-rich protein] |
-0.154 |
-0.515 |
| 65 |
245547_at |
AT4G15300
|
CYP702A2, cytochrome P450, family 702, subfamily A, polypeptide 2 |
-0.153 |
-2.010 |
| 66 |
262857_at |
AT1G14930
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.149 |
-0.515 |
| 67 |
254782_at |
AT4G12860
|
UNE14, unfertilized embryo sac 14 |
-0.148 |
-0.515 |