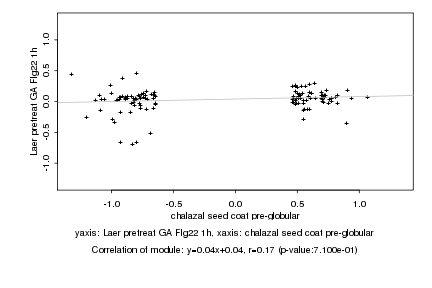
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
chalazal seed coat pre-globular |
Log2 signal ratio
Laer pretreat GA Flg22 1h |
| 1 |
262140_at |
AT1G52470
|
[alpha/beta-Hydrolases superfamily protein] |
1.064 |
0.073 |
| 2 |
263465_at |
AT2G31940
|
unknown |
0.937 |
0.054 |
| 3 |
254761_at |
AT4G13195
|
CLE44, CLAVATA3/ESR-RELATED 44 |
0.904 |
0.188 |
| 4 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
0.891 |
-0.354 |
| 5 |
258004_at |
AT3G29020
|
AtMYB110, myb domain protein 110, MYB110, myb domain protein 110 |
0.823 |
0.110 |
| 6 |
256011_at |
AT1G19230
|
[Riboflavin synthase-like superfamily protein] |
0.822 |
-0.029 |
| 7 |
265448_at |
AT2G46590
|
Dof-type zinc finger DNA-binding family protein |
0.807 |
0.081 |
| 8 |
245473_at |
AT4G16050
|
[Aminotransferase-like, plant mobile domain family protein] |
0.775 |
0.064 |
| 9 |
247022_at |
AT5G67050
|
[alpha/beta-Hydrolases superfamily protein] |
0.770 |
0.007 |
| 10 |
248254_at |
AT5G53320
|
[Leucine-rich repeat protein kinase family protein] |
0.768 |
0.056 |
| 11 |
252677_at |
AT3G44320
|
AtNIT3, NITRILASE 3, NIT3, nitrilase 3 |
0.759 |
0.041 |
| 12 |
247041_at |
AT5G67180
|
TOE3, target of early activation tagged (EAT) 3 |
0.744 |
-0.018 |
| 13 |
257517_at |
AT3G16330
|
unknown |
0.734 |
0.186 |
| 14 |
254294_at |
AT4G23070
|
ATRBL7, RHOMBOID-like protein 7, RBL7, RHOMBOID-like protein 7 |
0.726 |
0.101 |
| 15 |
267192_at |
AT2G30890
|
[Cytochrome b561/ferric reductase transmembrane protein family] |
0.718 |
0.102 |
| 16 |
262628_at |
AT1G06490
|
ATGSL07, Arabidopsis thaliana glucan synthase-like 7, atgsl7, CalS7, callose synthase 7, gsl07, GSL07, glucan synthase-like 7, GSL7, glucan synthase-like 7 |
0.713 |
0.079 |
| 17 |
250004_at |
AT5G18750
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.711 |
0.095 |
| 18 |
258326_at |
AT3G22760
|
SOL1 |
0.706 |
-0.006 |
| 19 |
260460_at |
AT1G68230
|
[Reticulon family protein] |
0.701 |
0.036 |
| 20 |
264204_at |
AT1G22710
|
ATSUC2, ARABIDOPSIS THALIANA SUCROSE-PROTON SYMPORTER 2, SUC2, sucrose-proton symporter 2, SUT1, SUCROSE TRANSPORTER 1 |
0.701 |
0.099 |
| 21 |
261893_at |
AT1G80690
|
[PPPDE putative thiol peptidase family protein] |
0.697 |
0.003 |
| 22 |
266216_at |
AT2G28810
|
[Dof-type zinc finger DNA-binding family protein] |
0.691 |
0.054 |
| 23 |
259252_at |
AT3G07610
|
IBM1, increase in bonsai methylation 1 |
0.691 |
0.104 |
| 24 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.689 |
0.156 |
| 25 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
0.643 |
0.053 |
| 26 |
267493_at |
AT2G30400
|
ATOFP2, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 2, OFP2, ovate family protein 2 |
0.638 |
0.301 |
| 27 |
250494_at |
AT5G09740
|
HAC11, HISTONE ACETYLTRANSFERASE OF THE CBP FAMILY 11, HAG05, HAG5, HISTONE ACETYLTRANSFERASE OF THE GNAT/MYST SUPERFAMILY 5, HAM2, histone acetyltransferase of the MYST family 2 |
0.613 |
0.140 |
| 28 |
246544_at |
AT5G15100
|
ATPIN8, PIN-FORMED 8, PIN8, PIN-FORMED 8 |
0.610 |
0.137 |
| 29 |
254411_at |
AT4G21420
|
[gypsy-like retrotransposon family, has a 6.9e-06 P-value blast match to GB:BAA84458 GAG-POL precursor (gypsy_Ty3-element) (Oryza sativa)gi|5902445|dbj|BAA84458.1| GAG-POL precursor (Oryza sativa (japonica cultivar-group)) (RIRE2) (Gypsy_Ty3-family)] |
0.601 |
0.060 |
| 30 |
250265_at |
AT5G12900
|
unknown |
0.598 |
-0.128 |
| 31 |
256989_at |
AT3G28580
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.596 |
0.292 |
| 32 |
247707_at |
AT5G59450
|
[GRAS family transcription factor] |
0.594 |
0.148 |
| 33 |
262114_at |
AT1G02860
|
BAH1, BENZOIC ACID HYPERSENSITIVE 1, NLA, nitrogen limitation adaptation |
0.585 |
0.090 |
| 34 |
260419_at |
AT1G69730
|
[Wall-associated kinase family protein] |
0.581 |
-0.116 |
| 35 |
263877_at |
AT2G21780
|
unknown |
0.568 |
0.029 |
| 36 |
250024_at |
AT5G18270
|
ANAC087, Arabidopsis NAC domain containing protein 87 |
0.558 |
0.254 |
| 37 |
253246_at |
AT4G34600
|
unknown |
0.552 |
-0.118 |
| 38 |
245130_at |
AT2G45340
|
[Leucine-rich repeat protein kinase family protein] |
0.546 |
-0.289 |
| 39 |
254174_at |
AT4G24120
|
ATYSL1, YELLOW STRIPE LIKE 1, YSL1, YELLOW STRIPE like 1 |
0.545 |
-0.131 |
| 40 |
262782_at |
AT1G13195
|
[RING/U-box superfamily protein] |
0.543 |
0.017 |
| 41 |
252457_at |
AT3G47180
|
[RING/U-box superfamily protein] |
0.542 |
-0.024 |
| 42 |
261113_at |
AT1G75400
|
[RING/U-box superfamily protein] |
0.536 |
0.133 |
| 43 |
247200_at |
AT5G65120
|
unknown |
0.527 |
0.247 |
| 44 |
257298_at |
AT3G28155
|
unknown |
0.521 |
0.054 |
| 45 |
255831_at |
AT2G33350
|
[CCT motif family protein] |
0.520 |
0.112 |
| 46 |
261100_at |
AT1G63020
|
NRPD1, NRPD1A, nuclear RNA polymerase D1A, POL IVA, NUCLEAR RNA POLYMERASE D 1A, SDE4, SMD2, SILENCING MOVEMENT DEFICIENT 2 |
0.512 |
0.130 |
| 47 |
249669_at |
AT5G35880
|
unknown |
0.507 |
0.065 |
| 48 |
259674_at |
AT1G77700
|
[Pathogenesis-related thaumatin superfamily protein] |
0.506 |
-0.017 |
| 49 |
266242_at |
AT2G27790
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.503 |
0.097 |
| 50 |
246401_at |
AT1G57560
|
AtMYB50, myb domain protein 50, MYB50, myb domain protein 50 |
0.498 |
0.143 |
| 51 |
250130_at |
AT5G16510
|
RGP5, reversibly glycosylated polypeptide 5, RGP5, reversibly glycosylated protein 5 |
0.495 |
0.231 |
| 52 |
266730_at |
AT2G03110
|
[RNA binding] |
0.487 |
-0.016 |
| 53 |
248469_at |
AT5G50820
|
anac097, NAC domain containing protein 97, NAC097, NAC domain containing protein 97 |
0.486 |
0.061 |
| 54 |
251923_at |
AT3G53880
|
AKR4C11, Aldo-keto reductase family 4 member C11 |
0.485 |
0.254 |
| 55 |
251262_at |
AT3G62080
|
[SNF7 family protein] |
0.484 |
0.265 |
| 56 |
252248_at |
AT3G49750
|
AtRLP44, receptor like protein 44, RLP44, receptor like protein 44 |
0.481 |
0.169 |
| 57 |
254258_at |
AT4G23410
|
TET5, tetraspanin5 |
0.480 |
0.026 |
| 58 |
263688_at |
AT1G26920
|
unknown |
0.478 |
-0.033 |
| 59 |
247640_at |
AT5G60610
|
[F-box/RNI-like superfamily protein] |
0.464 |
0.065 |
| 60 |
266425_at |
AT2G41300
|
SSL1, strictosidine synthase-like 1 |
0.463 |
-0.015 |
| 61 |
250724_at |
AT5G06330
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.461 |
0.089 |
| 62 |
258377_at |
AT3G17690
|
ATCNGC19, CYCLIC NUCLEOTIDE-GATED CHANNEL 19, CNGC19, cyclic nucleotide gated channel 19 |
0.460 |
0.251 |
| 63 |
248821_at |
AT5G47070
|
[Protein kinase superfamily protein] |
0.459 |
-0.015 |
| 64 |
254888_at |
AT4G11780
|
TRM10, TON1 Recruiting Motif 10 |
0.459 |
0.047 |
| 65 |
259809_at |
AT1G49800
|
unknown |
-1.330 |
0.454 |
| 66 |
247450_at |
AT5G62350
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-1.208 |
-0.252 |
| 67 |
252500_at |
AT3G46860
|
[Serine protease inhibitor, potato inhibitor I-type family protein] |
-1.133 |
0.022 |
| 68 |
267218_at |
AT2G02515
|
unknown |
-1.103 |
0.111 |
| 69 |
267169_at |
AT2G37540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-1.094 |
-0.135 |
| 70 |
261848_at |
AT1G11590
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-1.086 |
0.039 |
| 71 |
254295_at |
AT4G23080
|
unknown |
-1.059 |
0.048 |
| 72 |
251673_at |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
-1.012 |
0.266 |
| 73 |
246687_at |
AT5G33370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-1.006 |
0.141 |
| 74 |
266865_at |
AT2G29980
|
AtFAD3, FAD3, fatty acid desaturase 3 |
-1.001 |
-0.279 |
| 75 |
254122_at |
AT4G24510
|
CER2, ECERIFERUM 2, VC-2, VC2 |
-0.978 |
-0.330 |
| 76 |
250731_at |
AT5G06500
|
AGL96, AGAMOUS-like 96 |
-0.965 |
0.029 |
| 77 |
247785_at |
AT5G58820
|
[Subtilisin-like serine endopeptidase family protein] |
-0.953 |
0.025 |
| 78 |
249671_at |
AT5G35900
|
LBD35, LOB domain-containing protein 35 |
-0.943 |
0.068 |
| 79 |
257889_at |
AT3G17080
|
[Plant self-incompatibility protein S1 family] |
-0.939 |
0.047 |
| 80 |
261420_at |
AT1G07720
|
KCS3, 3-ketoacyl-CoA synthase 3 |
-0.934 |
-0.658 |
| 81 |
264390_at |
AT1G11950
|
[Transcription factor jumonji (jmjC) domain-containing protein] |
-0.931 |
-0.165 |
| 82 |
254489_at |
AT4G20800
|
[FAD-binding Berberine family protein] |
-0.929 |
0.069 |
| 83 |
255956_at |
AT1G22015
|
DD46 |
-0.920 |
0.090 |
| 84 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
-0.916 |
0.382 |
| 85 |
259786_at |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.893 |
0.055 |
| 86 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
-0.892 |
0.073 |
| 87 |
255019_at |
AT4G10210
|
unknown |
-0.888 |
0.034 |
| 88 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.883 |
0.060 |
| 89 |
246890_at |
AT5G25410
|
unknown |
-0.873 |
0.054 |
| 90 |
247526_at |
AT5G61470
|
[C2H2-like zinc finger protein] |
-0.873 |
0.093 |
| 91 |
261727_at |
AT1G76090
|
SMT3, sterol methyltransferase 3 |
-0.849 |
-0.166 |
| 92 |
251627_at |
AT3G57270
|
BG1, beta-1,3-glucanase 1 |
-0.847 |
0.097 |
| 93 |
255896_at |
AT1G17800
|
AtENODL22, ENODL22, early nodulin-like protein 22 |
-0.847 |
-0.025 |
| 94 |
251899_at |
AT3G54400
|
[Eukaryotic aspartyl protease family protein] |
-0.837 |
-0.685 |
| 95 |
250859_at |
AT5G04660
|
CYP77A4, cytochrome P450, family 77, subfamily A, polypeptide 4 |
-0.831 |
0.051 |
| 96 |
255357_at |
AT4G03930
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.828 |
-0.013 |
| 97 |
265590_at |
AT2G20160
|
ASK17, ARABIDOPSIS SKP1-LIKE 17, MEO, MEIDOS |
-0.822 |
0.030 |
| 98 |
252886_at |
AT4G39350
|
ATCESA2, ATH-A, CESA2, cellulose synthase A2 |
-0.817 |
-0.051 |
| 99 |
267392_at |
AT2G44490
|
BGLU26, BETA GLUCOSIDASE 26, PEN2, PENETRATION 2 |
-0.812 |
0.056 |
| 100 |
263431_at |
AT2G22170
|
PLAT2, PLAT domain protein 2 |
-0.807 |
-0.661 |
| 101 |
246978_at |
AT5G24910
|
CYP714A1, cytochrome P450, family 714, subfamily A, polypeptide 1, ELA1, EUI-like p450 A1 |
-0.806 |
0.455 |
| 102 |
259813_at |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
-0.800 |
0.038 |
| 103 |
256099_at |
AT1G13710
|
CYP78A5, cytochrome P450, family 78, subfamily A, polypeptide 5, KLU, KLUH |
-0.783 |
0.110 |
| 104 |
261203_at |
AT1G12845
|
unknown |
-0.779 |
-0.019 |
| 105 |
253459_at |
AT4G32080
|
unknown |
-0.775 |
0.096 |
| 106 |
258016_at |
AT3G19350
|
MPC, maternally expressed pab C-terminal |
-0.773 |
0.056 |
| 107 |
256310_at |
AT1G30360
|
ERD4, early-responsive to dehydration 4 |
-0.773 |
-0.101 |
| 108 |
249500_at |
AT5G39260
|
ATEXP21, ATEXPA21, expansin A21, ATHEXP ALPHA 1.20, EXP21, EXPANSIN 21, EXPA21, expansin A21 |
-0.771 |
0.051 |
| 109 |
251012_at |
AT5G02580
|
unknown |
-0.768 |
-0.051 |
| 110 |
246682_at |
AT5G33290
|
XGD1, xylogalacturonan deficient 1 |
-0.765 |
0.125 |
| 111 |
262797_at |
AT1G20840
|
AtTMT1, TMT1, tonoplast monosaccharide transporter1 |
-0.750 |
0.073 |
| 112 |
258744_at |
AT3G05830
|
unknown |
-0.747 |
0.138 |
| 113 |
267267_at |
AT2G02580
|
CYP71B9, cytochrome P450, family 71, subfamily B, polypeptide 9 |
-0.730 |
0.054 |
| 114 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
-0.728 |
0.101 |
| 115 |
263979_at |
AT2G42840
|
PDF1, protodermal factor 1 |
-0.720 |
0.177 |
| 116 |
247320_at |
AT5G64040
|
PSAN |
-0.719 |
-0.117 |
| 117 |
246624_at |
AT1G48910
|
YUC10, YUCCA 10 |
-0.714 |
0.034 |
| 118 |
256489_at |
AT1G31550
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.692 |
-0.504 |
| 119 |
255954_at |
AT1G22090
|
emb2204, embryo defective 2204 |
-0.686 |
0.130 |
| 120 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
-0.666 |
0.130 |
| 121 |
256309_at |
AT1G30380
|
PSAK, photosystem I subunit K |
-0.665 |
-0.107 |
| 122 |
267198_at |
AT2G30810
|
[Gibberellin-regulated family protein] |
-0.662 |
0.062 |
| 123 |
248202_at |
AT5G54220
|
[Cysteine-rich protein] |
-0.660 |
0.110 |
| 124 |
262455_at |
AT1G11310
|
ATMLO2, MILDEW RESISTANCE LOCUS O 2, MLO2, MILDEW RESISTANCE LOCUS O 2, PMR2, POWDERY MILDEW RESISTANT 2 |
-0.660 |
0.148 |
| 125 |
249450_at |
AT5G39440
|
SnRK1.3, SNF1-related protein kinase 1.3 |
-0.653 |
0.088 |
| 126 |
255644_at |
AT4G00870
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.651 |
-0.019 |
| 127 |
257769_at |
AT3G23050
|
AXR2, AUXIN RESISTANT 2, IAA7, indole-3-acetic acid 7 |
-0.650 |
-0.036 |