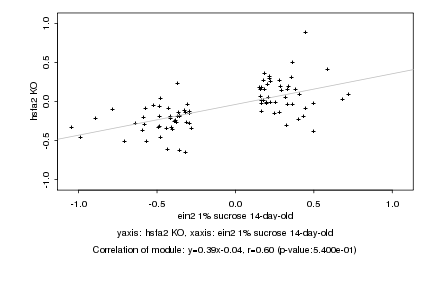
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ein2 1% sucrose 14-day-old |
Log2 signal ratio
hsfa2 KO |
| 1 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
0.719 |
0.099 |
| 2 |
260150_at |
AT1G52820
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.677 |
0.038 |
| 3 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
0.581 |
0.414 |
| 4 |
247467_at |
AT5G62130
|
[Per1-like family protein] |
0.497 |
-0.377 |
| 5 |
267626_at |
AT2G42250
|
CYP712A1, cytochrome P450, family 712, subfamily A, polypeptide 1 |
0.495 |
-0.021 |
| 6 |
265922_at |
AT2G18480
|
[Major facilitator superfamily protein] |
0.446 |
-0.088 |
| 7 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
0.441 |
0.891 |
| 8 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
0.430 |
-0.190 |
| 9 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
0.407 |
0.101 |
| 10 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
0.398 |
-0.219 |
| 11 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
0.381 |
0.159 |
| 12 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
0.363 |
-0.026 |
| 13 |
252618_at |
AT3G45140
|
ATLOX2, ARABIODOPSIS THALIANA LIPOXYGENASE 2, LOX2, lipoxygenase 2 |
0.362 |
0.506 |
| 14 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.355 |
0.320 |
| 15 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
0.334 |
0.196 |
| 16 |
261407_at |
AT1G18810
|
[phytochrome kinase substrate-related] |
0.330 |
-0.027 |
| 17 |
255786_at |
AT1G19670
|
ATCLH1, chlorophyllase 1, ATHCOR1, CORONATINE-INDUCED PROTEIN 1, CLH1, chlorophyllase 1, CORI1, CORONATINE-INDUCED PROTEIN 1 |
0.328 |
0.159 |
| 18 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
0.320 |
-0.298 |
| 19 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.314 |
0.061 |
| 20 |
265305_at |
AT2G20340
|
AAS, aromatic aldehyde synthase, AtAAS |
0.288 |
0.147 |
| 21 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
0.281 |
0.199 |
| 22 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
0.279 |
0.273 |
| 23 |
247055_at |
AT5G66740
|
unknown |
0.276 |
-0.132 |
| 24 |
250688_at |
AT5G06510
|
NF-YA10, nuclear factor Y, subunit A10 |
0.255 |
-0.010 |
| 25 |
263002_at |
AT1G54200
|
unknown |
0.245 |
-0.148 |
| 26 |
259560_at |
AT1G21270
|
WAK2, wall-associated kinase 2 |
0.220 |
0.262 |
| 27 |
256617_at |
AT3G22240
|
unknown |
0.219 |
-0.012 |
| 28 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
0.215 |
0.328 |
| 29 |
259609_at |
AT1G52410
|
AtTSA1, TSA1, TSK-associating protein 1 |
0.214 |
0.307 |
| 30 |
250651_at |
AT5G06900
|
CYP93D1, cytochrome P450, family 93, subfamily D, polypeptide 1 |
0.206 |
0.056 |
| 31 |
265147_at |
AT1G51380
|
[DEA(D/H)-box RNA helicase family protein] |
0.200 |
0.230 |
| 32 |
259346_at |
AT3G03910
|
glutamate dehydrogenase 3 |
0.196 |
-0.009 |
| 33 |
254693_at |
AT4G17880
|
MYC4 |
0.194 |
-0.022 |
| 34 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
0.183 |
0.160 |
| 35 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.182 |
0.368 |
| 36 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.176 |
0.025 |
| 37 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
0.173 |
0.274 |
| 38 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
0.166 |
0.185 |
| 39 |
254683_at |
AT4G13800
|
unknown |
0.163 |
-0.121 |
| 40 |
262213_at |
AT1G74870
|
[RING/U-box superfamily protein] |
0.163 |
0.019 |
| 41 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.161 |
-0.019 |
| 42 |
246235_at |
AT4G36830
|
HOS3-1 |
0.157 |
0.074 |
| 43 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
0.157 |
0.166 |
| 44 |
249208_at |
AT5G42650
|
AOS, allene oxide synthase, CYP74A, CYTOCHROME P450 74A, DDE2, DELAYED DEHISCENCE 2 |
0.152 |
0.182 |
| 45 |
248509_at |
AT5G50335
|
unknown |
-1.050 |
-0.320 |
| 46 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
-0.989 |
-0.448 |
| 47 |
247878_at |
AT5G57760
|
unknown |
-0.896 |
-0.215 |
| 48 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
-0.784 |
-0.091 |
| 49 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
-0.709 |
-0.511 |
| 50 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
-0.642 |
-0.269 |
| 51 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
-0.598 |
-0.362 |
| 52 |
263653_at |
AT1G04310
|
ERS2, ethylene response sensor 2 |
-0.590 |
-0.204 |
| 53 |
246935_at |
AT5G25350
|
EBF2, EIN3-binding F box protein 2 |
-0.586 |
-0.292 |
| 54 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.576 |
-0.083 |
| 55 |
248208_at |
AT5G53980
|
ATHB52, homeobox protein 52, HB52, homeobox protein 52 |
-0.572 |
-0.503 |
| 56 |
257636_at |
AT3G26200
|
CYP71B22, cytochrome P450, family 71, subfamily B, polypeptide 22 |
-0.528 |
-0.041 |
| 57 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.493 |
-0.325 |
| 58 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
-0.490 |
-0.317 |
| 59 |
252764_at |
AT3G42790
|
AL3, alfin-like 3 |
-0.487 |
-0.058 |
| 60 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
-0.486 |
-0.179 |
| 61 |
247474_at |
AT5G62280
|
unknown |
-0.481 |
-0.449 |
| 62 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
-0.479 |
0.046 |
| 63 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.444 |
-0.334 |
| 64 |
248564_at |
AT5G49700
|
AHL17, AT-hook motif nuclear localized protein 17 |
-0.438 |
-0.607 |
| 65 |
248496_at |
AT5G50790
|
AtSWEET10, SWEET10 |
-0.427 |
-0.084 |
| 66 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
-0.420 |
-0.211 |
| 67 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
-0.418 |
-0.183 |
| 68 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
-0.409 |
-0.332 |
| 69 |
250049_at |
AT5G17780
|
[alpha/beta-Hydrolases superfamily protein] |
-0.404 |
-0.350 |
| 70 |
266383_at |
AT2G14580
|
ATPRB1, basic pathogenesis-related protein 1, PRB1, basic pathogenesis-related protein 1 |
-0.394 |
-0.253 |
| 71 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.383 |
-0.231 |
| 72 |
261327_at |
AT1G44830
|
[Integrase-type DNA-binding superfamily protein] |
-0.382 |
-0.264 |
| 73 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.372 |
0.237 |
| 74 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
-0.372 |
-0.183 |
| 75 |
258925_at |
AT3G10420
|
SPD1, SEEDLING PLASTID DEVELOPMENT 1 |
-0.365 |
-0.134 |
| 76 |
257701_at |
AT3G12710
|
[DNA glycosylase superfamily protein] |
-0.363 |
-0.181 |
| 77 |
258434_at |
AT3G16770
|
ATEBP, ethylene-responsive element binding protein, EBP, ethylene-responsive element binding protein, ERF72, ETHYLENE RESPONSE FACTOR 72, RAP2.3, RELATED TO AP2 3 |
-0.361 |
-0.172 |
| 78 |
253763_at |
AT4G28850
|
ATXTH26, XTH26, xyloglucan endotransglucosylase/hydrolase 26 |
-0.357 |
-0.625 |
| 79 |
264902_at |
AT1G23060
|
MDP40, MICROTUBULE DESTABILIZING PROTEIN 40 |
-0.327 |
-0.110 |
| 80 |
263890_at |
AT2G37030
|
SAUR46, SMALL AUXIN UPREGULATED RNA 46 |
-0.324 |
-0.645 |
| 81 |
249887_at |
AT5G22310
|
unknown |
-0.322 |
-0.133 |
| 82 |
252859_at |
AT4G39780
|
[Integrase-type DNA-binding superfamily protein] |
-0.314 |
-0.257 |
| 83 |
245098_at |
AT2G40940
|
ERS, ETHYLENE RESPONSE SENSOR, ERS1, ethylene response sensor 1 |
-0.310 |
-0.033 |
| 84 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
-0.299 |
-0.275 |
| 85 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
-0.294 |
-0.116 |
| 86 |
246195_at |
AT4G36410
|
UBC17, ubiquitin-conjugating enzyme 17 |
-0.294 |
-0.149 |
| 87 |
253437_at |
AT4G32460
|
unknown |
-0.285 |
-0.345 |