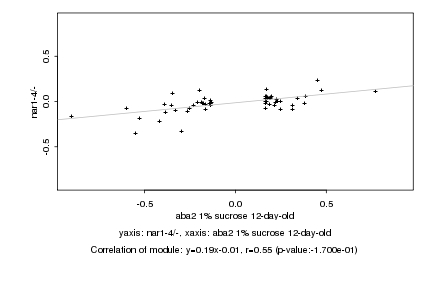
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
aba2 1% sucrose 12-day-old |
Log2 signal ratio
nar1-4/- |
| 1 |
264872_at |
AT1G24260
|
AGL9, AGAMOUS-like 9, SEP3, SEPALLATA3 |
0.770 |
0.111 |
| 2 |
248371_at |
AT5G51810
|
AT2353, ATGA20OX2, GA20OX2, gibberellin 20 oxidase 2 |
0.473 |
0.122 |
| 3 |
266658_at |
AT2G25735
|
unknown |
0.448 |
0.238 |
| 4 |
265327_at |
AT2G18210
|
unknown |
0.384 |
0.065 |
| 5 |
257536_at |
AT3G02800
|
AtPFA-DSP3, PFA-DSP3, plant and fungi atypical dual-specificity phosphatase 3 |
0.375 |
-0.013 |
| 6 |
259211_at |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
0.336 |
0.041 |
| 7 |
259372_at |
AT1G69120
|
AGL7, AGAMOUS-like 7, AP1, APETALA1, AtAP1 |
0.311 |
-0.081 |
| 8 |
267254_at |
AT2G23030
|
SNRK2-9, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-9, SNRK2.9, SNF1-related protein kinase 2.9 |
0.309 |
-0.038 |
| 9 |
246018_at |
AT5G10695
|
unknown |
0.247 |
0.003 |
| 10 |
248528_at |
AT5G50760
|
SAUR55, SMALL AUXIN UPREGULATED RNA 55 |
0.243 |
-0.079 |
| 11 |
264202_at |
AT1G22810
|
[Integrase-type DNA-binding superfamily protein] |
0.228 |
0.009 |
| 12 |
247753_at |
AT5G59070
|
[UDP-Glycosyltransferase superfamily protein] |
0.222 |
0.023 |
| 13 |
265740_at |
AT2G01150
|
RHA2B, RING-H2 finger protein 2B |
0.220 |
-0.008 |
| 14 |
246532_at |
AT5G15870
|
[glycosyl hydrolase family 81 protein] |
0.214 |
-0.042 |
| 15 |
250107_at |
AT5G15330
|
ATSPX4, ARABIDOPSIS THALIANA SPX DOMAIN GENE 4, SPX4, SPX domain gene 4 |
0.194 |
0.063 |
| 16 |
251012_at |
AT5G02580
|
unknown |
0.190 |
0.043 |
| 17 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
0.188 |
0.051 |
| 18 |
261506_at |
AT1G71697
|
ATCK1, choline kinase 1, CK, CHOLINE KINASE, CK1, choline kinase 1 |
0.184 |
0.039 |
| 19 |
253696_at |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
0.182 |
-0.023 |
| 20 |
254921_at |
AT4G11300
|
unknown |
0.176 |
0.034 |
| 21 |
258194_at |
AT3G29170
|
unknown |
0.172 |
0.047 |
| 22 |
259137_at |
AT3G10300
|
[Calcium-binding EF-hand family protein] |
0.168 |
0.007 |
| 23 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
0.168 |
0.137 |
| 24 |
245768_at |
AT1G33590
|
[Leucine-rich repeat (LRR) family protein] |
0.168 |
0.062 |
| 25 |
254571_at |
AT4G19370
|
unknown |
0.164 |
0.008 |
| 26 |
265955_at |
AT2G37280
|
ABCG33, ATP-binding cassette G33, ATPDR5, PLEIOTROPIC DRUG RESISTANCE 5, PDR5, pleiotropic drug resistance 5 |
0.163 |
0.036 |
| 27 |
247043_at |
AT5G66880
|
SNRK2-3, SUCROSE NONFERMENTING 1 (SNF1)-RELATED PROTEIN KINASE 2-3, SNRK2.3, sucrose nonfermenting 1(SNF1)-related protein kinase 2.3, SRK2I |
0.162 |
-0.011 |
| 28 |
259312_at |
AT3G05200
|
ATL6, Arabidopsis toxicos en levadura 6 |
0.162 |
0.057 |
| 29 |
256818_at |
AT3G21420
|
LBO1, LATERAL BRANCHING OXIDOREDUCTASE 1 |
0.162 |
-0.073 |
| 30 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
-0.906 |
-0.159 |
| 31 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.601 |
-0.068 |
| 32 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
-0.550 |
-0.344 |
| 33 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
-0.530 |
-0.187 |
| 34 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.422 |
-0.212 |
| 35 |
252618_at |
AT3G45140
|
ATLOX2, ARABIODOPSIS THALIANA LIPOXYGENASE 2, LOX2, lipoxygenase 2 |
-0.395 |
-0.026 |
| 36 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
-0.388 |
-0.111 |
| 37 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
-0.352 |
-0.038 |
| 38 |
262357_at |
AT1G73040
|
[Mannose-binding lectin superfamily protein] |
-0.350 |
0.092 |
| 39 |
260357_at |
AT1G69260
|
AFP1, ABI five binding protein |
-0.333 |
-0.099 |
| 40 |
260353_at |
AT1G69230
|
SP1L2, SPIRAL1-like2 |
-0.300 |
-0.327 |
| 41 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
-0.268 |
-0.109 |
| 42 |
267569_at |
AT2G30790
|
PSBP-2, photosystem II subunit P-2 |
-0.254 |
-0.072 |
| 43 |
248006_at |
AT5G56250
|
HAP8, HAPLESS 8 |
-0.236 |
-0.041 |
| 44 |
250002_at |
AT5G18690
|
AGP25, arabinogalactan protein 25, ATAGP25, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEINS 25 |
-0.211 |
0.000 |
| 45 |
246260_at |
AT1G31820
|
PUT1, POLYAMINE UPTAKE TRANSPORTER 1 |
-0.203 |
0.127 |
| 46 |
264696_at |
AT1G70230
|
AXY4, ALTERED XYLOGLUCAN 4, TBL27, TRICHOME BIREFRINGENCE-LIKE 27 |
-0.191 |
-0.000 |
| 47 |
267423_at |
AT2G35060
|
KUP11, K+ uptake permease 11 |
-0.186 |
-0.013 |
| 48 |
254413_at |
AT4G21440
|
ATM4, A. THALIANA MYB 4, ATMYB102, MYB-like 102, MYB102, MYB102, MYB-like 102 |
-0.180 |
-0.020 |
| 49 |
262154_at |
AT1G52700
|
[alpha/beta-Hydrolases superfamily protein] |
-0.172 |
0.043 |
| 50 |
265350_at |
AT2G22620
|
[Rhamnogalacturonate lyase family protein] |
-0.167 |
-0.083 |
| 51 |
260144_at |
AT1G71960
|
ABCG25, ATP-binding casette G25, ATABCG25, Arabidopsis thaliana ATP-binding cassette G25 |
-0.165 |
-0.032 |
| 52 |
264054_at |
AT2G22540
|
AGL22, AGAMOUS-like 22, FAQ1, Flowering Arabidopsis QTL1, SVP, SHORT VEGETATIVE PHASE |
-0.143 |
-0.018 |
| 53 |
245127_at |
AT2G47600
|
ATMHX, magnesium/proton exchanger, ATMHX1, MHX, magnesium/proton exchanger, MHX1, MAGNESIUM/PROTON EXCHANGER 1 |
-0.142 |
-0.037 |
| 54 |
259334_at |
AT3G03790
|
[ankyrin repeat family protein / regulator of chromosome condensation (RCC1) family protein] |
-0.141 |
-0.020 |
| 55 |
254894_at |
AT4G11840
|
PLDGAMMA3, phospholipase D gamma 3 |
-0.139 |
0.017 |
| 56 |
245011_at |
ATCG00430
|
PSBG, photosystem II reaction center protein G |
-0.139 |
-0.012 |
| 57 |
245352_at |
AT4G15490
|
UGT84A3 |
-0.136 |
-0.008 |