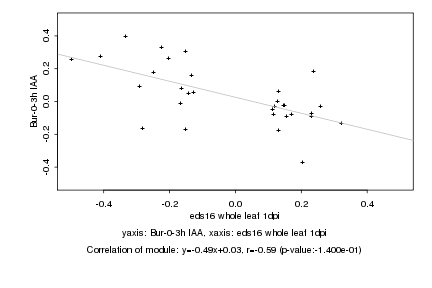
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
eds16 whole leaf 1dpi |
Log2 signal ratio
Bur-0-3h IAA |
| 1 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
0.322 |
-0.130 |
| 2 |
248867_at |
AT5G46830
|
ATNIG1, NACL-INDUCIBLE GENE 1, NIG1, NACL-inducible gene 1 |
0.256 |
-0.025 |
| 3 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
0.237 |
0.184 |
| 4 |
245465_at |
AT4G16590
|
ATCSLA01, cellulose synthase-like A01, ATCSLA1, CELLULOSE SYNTHASE-LIKE A1, CSLA01, CSLA01, cellulose synthase-like A01 |
0.229 |
-0.070 |
| 5 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
0.228 |
-0.090 |
| 6 |
260205_at |
AT1G70700
|
JAZ9, JASMONATE-ZIM-DOMAIN PROTEIN 9, TIFY7 |
0.203 |
-0.371 |
| 7 |
255401_at |
AT4G03600
|
unknown |
0.168 |
-0.076 |
| 8 |
264638_at |
AT1G65480
|
FT, FLOWERING LOCUS T, RSB8, REDUCED STEM BRANCHING 8 |
0.152 |
-0.091 |
| 9 |
251591_at |
AT3G57680
|
[Peptidase S41 family protein] |
0.148 |
-0.019 |
| 10 |
251598_at |
AT3G57600
|
[Integrase-type DNA-binding superfamily protein] |
0.144 |
-0.021 |
| 11 |
261851_at |
AT1G50460
|
ATHKL1, HKL1, hexokinase-like 1 |
0.130 |
-0.172 |
| 12 |
265300_at |
AT2G13950
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.129 |
0.066 |
| 13 |
260341_at |
AT1G69170
|
AtSPL6, SPL6, SQUAMOSA PROMOTER BINDING PROTEIN (SBP)-domain transcription factor 6 |
0.126 |
0.002 |
| 14 |
256509_at |
AT1G75300
|
[NmrA-like negative transcriptional regulator family protein] |
0.116 |
-0.025 |
| 15 |
257447_at |
AT2G04230
|
[FBD, F-box and Leucine Rich Repeat domains containing protein] |
0.114 |
-0.078 |
| 16 |
248553_at |
AT5G50170
|
[C2 calcium/lipid-binding and GRAM domain containing protein] |
0.112 |
-0.047 |
| 17 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
-0.500 |
0.259 |
| 18 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
-0.412 |
0.277 |
| 19 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
-0.336 |
0.397 |
| 20 |
262222_at |
AT1G74700
|
NUZ, TRZ1, tRNAse Z1 |
-0.294 |
0.096 |
| 21 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.285 |
-0.160 |
| 22 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
-0.249 |
0.177 |
| 23 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
-0.226 |
0.333 |
| 24 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
-0.204 |
0.263 |
| 25 |
251986_at |
AT3G53310
|
[AP2/B3-like transcriptional factor family protein] |
-0.168 |
-0.012 |
| 26 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
-0.164 |
0.084 |
| 27 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
-0.153 |
0.309 |
| 28 |
259671_at |
AT1G52290
|
AtPERK15, proline-rich extensin-like receptor kinase 15, PERK15, proline-rich extensin-like receptor kinase 15 |
-0.152 |
-0.165 |
| 29 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
-0.144 |
0.049 |
| 30 |
252345_at |
AT3G48640
|
unknown |
-0.136 |
0.163 |
| 31 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
-0.130 |
0.057 |