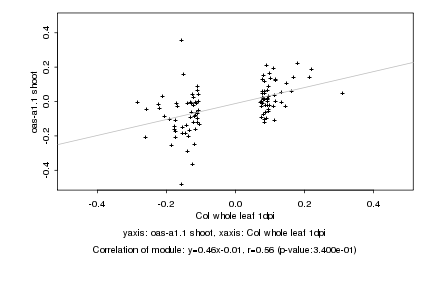
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col whole leaf 1dpi |
Log2 signal ratio
oas-a1.1 shoot |
| 1 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.310 |
0.049 |
| 2 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
0.220 |
0.188 |
| 3 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.212 |
0.144 |
| 4 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
0.177 |
0.222 |
| 5 |
264638_at |
AT1G65480
|
FT, FLOWERING LOCUS T, RSB8, REDUCED STEM BRANCHING 8 |
0.168 |
0.144 |
| 6 |
254323_at |
AT4G22620
|
SAUR34, SMALL AUXIN UPREGULATED RNA 34 |
0.161 |
0.064 |
| 7 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
0.147 |
0.109 |
| 8 |
261924_at |
AT1G22550
|
[Major facilitator superfamily protein] |
0.145 |
-0.027 |
| 9 |
245912_at |
AT5G19600
|
SULTR3;5, sulfate transporter 3;5 |
0.132 |
0.058 |
| 10 |
251317_at |
AT3G61490
|
[Pectin lyase-like superfamily protein] |
0.131 |
-0.004 |
| 11 |
263436_at |
AT2G28690
|
unknown |
0.116 |
0.124 |
| 12 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
0.116 |
0.004 |
| 13 |
247511_at |
AT5G62040
|
BFT, brother of FT and TFL1 |
0.114 |
0.132 |
| 14 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.113 |
-0.110 |
| 15 |
250881_at |
AT5G04080
|
unknown |
0.111 |
0.040 |
| 16 |
254993_at |
AT4G10730
|
[Protein kinase superfamily protein] |
0.110 |
-0.025 |
| 17 |
257024_at |
AT3G19100
|
[Protein kinase superfamily protein] |
0.108 |
0.193 |
| 18 |
261247_at |
AT1G20070
|
unknown |
0.100 |
0.138 |
| 19 |
263465_at |
AT2G31940
|
unknown |
0.096 |
-0.022 |
| 20 |
264053_at |
AT2G22560
|
NET2D, Networked 2D |
0.096 |
0.165 |
| 21 |
247640_at |
AT5G60610
|
[F-box/RNI-like superfamily protein] |
0.095 |
-0.041 |
| 22 |
255921_at |
AT1G22240
|
APUM8, pumilio 8, PUM8, pumilio 8 |
0.094 |
0.089 |
| 23 |
247351_at |
AT5G63790
|
ANAC102, NAC domain containing protein 102, NAC102, NAC domain containing protein 102 |
0.093 |
-0.056 |
| 24 |
255289_at |
AT4G04690
|
[F-box and associated interaction domains-containing protein] |
0.093 |
0.035 |
| 25 |
263179_at |
AT1G05710
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.091 |
0.067 |
| 26 |
265634_at |
AT2G25530
|
[AFG1-like ATPase family protein] |
0.090 |
0.017 |
| 27 |
263124_at |
AT1G78480
|
[Prenyltransferase family protein] |
0.090 |
0.003 |
| 28 |
264741_at |
AT1G62290
|
[Saposin-like aspartyl protease family protein] |
0.090 |
0.018 |
| 29 |
252005_at |
AT3G52810
|
ATPAP21, PURPLE ACID PHOSPHATASE 21, PAP21, purple acid phosphatase 21 |
0.090 |
-0.019 |
| 30 |
254939_at |
AT4G10800
|
unknown |
0.089 |
-0.095 |
| 31 |
245724_at |
AT1G73390
|
[Endosomal targeting BRO1-like domain-containing protein] |
0.088 |
0.210 |
| 32 |
254304_at |
AT4G22270
|
ATMRB1, ARABIDOPSIS MEMBRANE RELATED BIGGER1, MRB1, MEMBRANE RELATED BIGGER1 |
0.085 |
-0.061 |
| 33 |
258603_at |
AT3G02990
|
ATHSFA1E, heat shock transcription factor A1E, HSFA1E, heat shock transcription factor A1E |
0.085 |
-0.020 |
| 34 |
247965_at |
AT5G56540
|
AGP14, arabinogalactan protein 14, ATAGP14 |
0.084 |
-0.099 |
| 35 |
251825_at |
AT3G55100
|
ABCG17, ATP-binding cassette G17 |
0.084 |
0.052 |
| 36 |
260080_at |
AT1G78160
|
APUM7, pumilio 7, PUM7, pumilio 7 |
0.083 |
0.014 |
| 37 |
264907_at |
AT2G17280
|
[Phosphoglycerate mutase family protein] |
0.083 |
0.061 |
| 38 |
264186_at |
AT1G54570
|
PES1, phytyl ester synthase 1 |
0.082 |
0.121 |
| 39 |
257956_at |
AT3G25400
|
unknown |
0.082 |
-0.117 |
| 40 |
258938_at |
AT3G10080
|
[RmlC-like cupins superfamily protein] |
0.080 |
0.013 |
| 41 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.080 |
0.154 |
| 42 |
245680_at |
AT1G56570
|
PGN, PENTATRICOPEPTIDE REPEAT PROTEIN FOR GERMINATION ON NaCl |
0.079 |
-0.075 |
| 43 |
246492_at |
AT5G16140
|
[Peptidyl-tRNA hydrolase family protein] |
0.079 |
0.018 |
| 44 |
267327_at |
AT2G19410
|
[U-box domain-containing protein kinase family protein] |
0.078 |
0.064 |
| 45 |
259704_at |
AT1G77680
|
[Ribonuclease II/R family protein] |
0.078 |
-0.011 |
| 46 |
245467_at |
AT4G16610
|
[C2H2-like zinc finger protein] |
0.076 |
0.049 |
| 47 |
262478_at |
AT1G11170
|
unknown |
0.076 |
0.029 |
| 48 |
265484_at |
AT2G15820
|
OTP51, ORGANELLE TRANSCRIPT PROCESSING 51 |
0.076 |
0.131 |
| 49 |
245225_at |
AT3G29800
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.075 |
-0.024 |
| 50 |
265544_at |
AT2G28260
|
ATCNGC15, cyclic nucleotide-gated channel 15, CNGC15, cyclic nucleotide-gated channel 15 |
0.075 |
-0.091 |
| 51 |
265928_at |
AT2G18580
|
[hAT-like transposase family (hobo/Ac/Tam3), has a 3.7e-98 P-value blast match to GB:AAD24567 transposase Tag2 (hAT-element) (Arabidopsis thaliana)] |
0.074 |
0.002 |
| 52 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.074 |
0.000 |
| 53 |
261171_at |
AT1G04880
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
0.072 |
-0.005 |
| 54 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
-0.284 |
-0.000 |
| 55 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
-0.263 |
-0.208 |
| 56 |
267546_at |
AT2G32680
|
AtRLP23, receptor like protein 23, RLP23, receptor like protein 23 |
-0.259 |
-0.042 |
| 57 |
264774_at |
AT1G22890
|
unknown |
-0.225 |
-0.015 |
| 58 |
247314_at |
AT5G64000
|
ATSAL2, SAL2 |
-0.220 |
-0.036 |
| 59 |
259012_at |
AT3G07360
|
ATPUB9, ARABIDOPSIS THALIANA PLANT U-BOX 9, PUB9, plant U-box 9 |
-0.213 |
0.031 |
| 60 |
249583_at |
AT5G37770
|
CML24, CALMODULIN-LIKE 24, TCH2, TOUCH 2 |
-0.207 |
-0.083 |
| 61 |
266658_at |
AT2G25735
|
unknown |
-0.192 |
-0.099 |
| 62 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
-0.186 |
-0.254 |
| 63 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
-0.178 |
-0.158 |
| 64 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
-0.177 |
-0.140 |
| 65 |
267289_at |
AT2G23770
|
LYK4, LysM-containing receptor-like kinase 4 |
-0.175 |
-0.209 |
| 66 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
-0.175 |
-0.171 |
| 67 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
-0.174 |
-0.107 |
| 68 |
266447_at |
AT2G43290
|
MSS3, multicopy suppressors of snf4 deficiency in yeast 3 |
-0.172 |
-0.011 |
| 69 |
259211_at |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
-0.168 |
-0.025 |
| 70 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
-0.157 |
0.359 |
| 71 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
-0.157 |
-0.477 |
| 72 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
-0.156 |
-0.181 |
| 73 |
261375_at |
AT1G53160
|
FTM6, FLORAL TRANSITION AT THE MERISTEM6, SPL4, squamosa promoter binding protein-like 4 |
-0.155 |
-0.147 |
| 74 |
250603_at |
AT5G07820
|
[Plant calmodulin-binding protein-related] |
-0.151 |
0.162 |
| 75 |
260472_at |
AT1G10990
|
unknown |
-0.146 |
-0.182 |
| 76 |
248646_at |
AT5G49100
|
unknown |
-0.143 |
-0.137 |
| 77 |
247685_at |
AT5G59680
|
[Leucine-rich repeat protein kinase family protein] |
-0.141 |
-0.288 |
| 78 |
250936_at |
AT5G03120
|
unknown |
-0.140 |
-0.007 |
| 79 |
263182_at |
AT1G05575
|
unknown |
-0.136 |
-0.199 |
| 80 |
256275_at |
AT3G12110
|
ACT11, actin-11 |
-0.135 |
-0.167 |
| 81 |
265893_at |
AT2G15042
|
[Leucine-rich repeat (LRR) family protein] |
-0.131 |
-0.088 |
| 82 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.131 |
-0.000 |
| 83 |
259841_at |
AT1G52200
|
[PLAC8 family protein] |
-0.130 |
-0.061 |
| 84 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
-0.128 |
-0.007 |
| 85 |
257139_at |
AT3G28890
|
AtRLP43, receptor like protein 43, RLP43, receptor like protein 43 |
-0.126 |
-0.364 |
| 86 |
248179_at |
AT5G54380
|
THE1, THESEUS1 |
-0.126 |
0.041 |
| 87 |
266371_at |
AT2G41410
|
[Calcium-binding EF-hand family protein] |
-0.124 |
-0.020 |
| 88 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
-0.124 |
-0.121 |
| 89 |
261949_at |
AT1G64670
|
BDG1, BODYGUARD1, CED1, 9-CIS EPOXYCAROTENOID DIOXYGENASE DEFECTIVE 1 |
-0.123 |
0.027 |
| 90 |
263914_at |
AT2G36400
|
AtGRF3, growth-regulating factor 3, GRF3, growth-regulating factor 3 |
-0.121 |
-0.082 |
| 91 |
247071_at |
AT5G66640
|
DAR3, DA1-related protein 3 |
-0.121 |
-0.250 |
| 92 |
261027_at |
AT1G01340
|
ACBK1, ATCNGC10, cyclic nucleotide gated channel 10, CNGC10, cyclic nucleotide gated channel 10 |
-0.118 |
-0.159 |
| 93 |
260143_at |
AT1G71880
|
ATSUC1, ARABIDOPSIS THALIANA SUCROSE-PROTON SYMPORTER 1, SUC1, sucrose-proton symporter 1 |
-0.117 |
-0.003 |
| 94 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
-0.116 |
-0.076 |
| 95 |
255912_at |
AT1G66960
|
LUP5 |
-0.115 |
-0.061 |
| 96 |
253776_at |
AT4G28390
|
AAC3, ADP/ATP carrier 3, ATAAC3 |
-0.113 |
-0.007 |
| 97 |
267083_at |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
-0.112 |
-0.118 |
| 98 |
257206_at |
AT3G16530
|
[Legume lectin family protein] |
-0.111 |
-0.068 |
| 99 |
261308_at |
AT1G48480
|
RKL1, receptor-like kinase 1 |
-0.111 |
-0.094 |
| 100 |
249409_at |
AT5G40340
|
[Tudor/PWWP/MBT superfamily protein] |
-0.110 |
0.069 |
| 101 |
254101_at |
AT4G25000
|
AMY1, alpha-amylase-like, ATAMY1 |
-0.110 |
0.091 |
| 102 |
250435_at |
AT5G10380
|
ATRING1, RING1 |
-0.109 |
-0.049 |
| 103 |
267423_at |
AT2G35060
|
KUP11, K+ uptake permease 11 |
-0.108 |
0.042 |
| 104 |
264574_at |
AT1G05300
|
ZIP5, zinc transporter 5 precursor |
-0.108 |
0.005 |
| 105 |
260239_at |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
-0.107 |
-0.128 |