|
probeID |
AGICode |
Annotation |
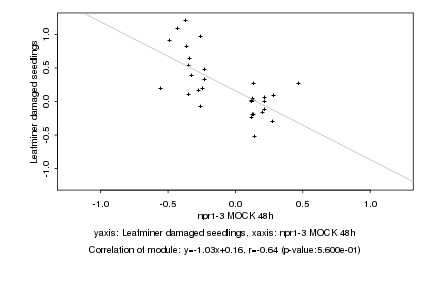
Log2 signal ratio
npr1-3 MOCK 48h |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
246888_at |
AT5G26270
|
unknown |
0.462 |
0.273 |
| 2 |
247224_at |
AT5G65080
|
AGL68, AGAMOUS-like 68, MAF5, MADS AFFECTING FLOWERING 5 |
0.281 |
0.099 |
| 3 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.271 |
-0.287 |
| 4 |
267564_at |
AT2G30740
|
[Protein kinase superfamily protein] |
0.215 |
0.069 |
| 5 |
265051_at |
AT1G52100
|
[Mannose-binding lectin superfamily protein] |
0.211 |
0.008 |
| 6 |
257551_at |
AT3G13270
|
unknown |
0.209 |
-0.115 |
| 7 |
254933_at |
AT4G11130
|
RDR2, RNA-dependent RNA polymerase 2, SMD1, SILENCING MOVEMENT DEFICIENT 1 |
0.195 |
-0.158 |
| 8 |
265726_at |
AT2G32010
|
CVL1, CVP2 like 1 |
0.139 |
-0.520 |
| 9 |
262549_at |
AT1G31290
|
AGO3, ARGONAUTE 3 |
0.134 |
0.022 |
| 10 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
0.130 |
0.273 |
| 11 |
259888_at |
AT1G76350
|
NLP5, NIN-like protein 5 |
0.129 |
-0.193 |
| 12 |
258043_at |
AT3G21290
|
[dentin sialophosphoprotein-related] |
0.120 |
-0.184 |
| 13 |
252232_at |
AT3G49760
|
AtbZIP5, basic leucine-zipper 5, bZIP5, basic leucine-zipper 5 |
0.120 |
0.059 |
| 14 |
246473_at |
AT5G17140
|
[Cysteine proteinases superfamily protein] |
0.117 |
0.002 |
| 15 |
263851_at |
AT2G04460
|
unknown |
0.117 |
-0.233 |
| 16 |
256970_at |
AT3G21090
|
ABCG15, ATP-binding cassette G15 |
0.117 |
0.017 |
| 17 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
-0.563 |
0.197 |
| 18 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
-0.494 |
0.915 |
| 19 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
-0.434 |
1.091 |
| 20 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
-0.378 |
1.222 |
| 21 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
-0.367 |
0.832 |
| 22 |
259764_at |
AT1G64280
|
ATNPR1, ARABIDOPSIS NONEXPRESSER OF PR GENES 1, NIM1, NON-INDUCIBLE IMMUNITY 1, NPR1, NONEXPRESSER OF PR GENES 1, SAI1, SALICYLIC ACID INSENSITIVE 1 |
-0.351 |
0.119 |
| 23 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
-0.349 |
0.545 |
| 24 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
-0.342 |
0.651 |
| 25 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
-0.332 |
0.395 |
| 26 |
250500_at |
AT5G09530
|
PELPK1, Pro-Glu-Leu|Ile|Val-Pro-Lys 1, PRP10, proline-rich protein 10 |
-0.281 |
0.178 |
| 27 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
-0.266 |
0.976 |
| 28 |
264987_at |
AT1G27030
|
unknown |
-0.260 |
-0.070 |
| 29 |
252060_at |
AT3G52430
|
ATPAD4, ARABIDOPSIS PHYTOALEXIN DEFICIENT 4, PAD4, PHYTOALEXIN DEFICIENT 4 |
-0.250 |
0.207 |
| 30 |
251895_at |
AT3G54420
|
ATCHITIV, CHITINASE CLASS IV, ATEP3, homolog of carrot EP3-3 chitinase, CHIV, EP3, homolog of carrot EP3-3 chitinase |
-0.237 |
0.484 |
| 31 |
249485_at |
AT5G39020
|
[Malectin/receptor-like protein kinase family protein] |
-0.232 |
0.329 |