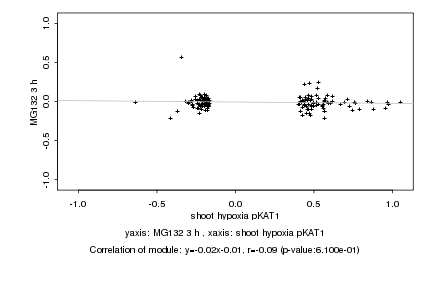
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
shoot hypoxia pKAT1 |
Log2 signal ratio
MG132 3 h |
| 1 |
252650_at |
AT3G44690
|
unknown |
1.049 |
-0.009 |
| 2 |
267089_at |
AT2G38300
|
[myb-like HTH transcriptional regulator family protein] |
0.969 |
-0.037 |
| 3 |
260672_at |
AT1G19480
|
[DNA glycosylase superfamily protein] |
0.966 |
-0.012 |
| 4 |
250650_at |
AT5G06850
|
FTIP1, FT-interacting protein 1 |
0.953 |
-0.078 |
| 5 |
259423_at |
AT1G13880
|
[ELM2 domain-containing protein] |
0.878 |
-0.094 |
| 6 |
257367_at |
AT2G25780
|
unknown |
0.861 |
-0.011 |
| 7 |
262680_at |
AT1G75880
|
[SGNH hydrolase-type esterase superfamily protein] |
0.840 |
0.005 |
| 8 |
261776_at |
AT1G76190
|
SAUR56, SMALL AUXIN UPREGULATED RNA 56 |
0.787 |
-0.095 |
| 9 |
246653_at |
AT5G35200
|
[ENTH/ANTH/VHS superfamily protein] |
0.761 |
-0.019 |
| 10 |
253963_at |
AT4G26470
|
[Calcium-binding EF-hand family protein] |
0.754 |
-0.012 |
| 11 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
0.742 |
-0.103 |
| 12 |
254188_at |
AT4G23920
|
ATUGE2, UDP-GLC 4-EPIMERASE 2, UGE2, UDP-D-glucose/UDP-D-galactose 4-epimerase 2 |
0.726 |
-0.059 |
| 13 |
264782_at |
AT1G08810
|
AtMYB60, myb domain protein 60, MYB60, myb domain protein 60 |
0.709 |
0.038 |
| 14 |
245385_at |
AT4G14020
|
[Rapid alkalinization factor (RALF) family protein] |
0.692 |
-0.002 |
| 15 |
254870_at |
AT4G11900
|
[S-locus lectin protein kinase family protein] |
0.664 |
-0.038 |
| 16 |
264437_at |
AT1G27510
|
EX2, EXECUTER 2 |
0.618 |
0.006 |
| 17 |
266600_at |
AT2G46070
|
ATMPK12, MAPK12, MPK12, mitogen-activated protein kinase 12 |
0.612 |
0.067 |
| 18 |
251454_at |
AT3G60080
|
[RING/U-box superfamily protein] |
0.603 |
-0.024 |
| 19 |
265222_at |
AT2G01970
|
[Endomembrane protein 70 protein family] |
0.592 |
-0.017 |
| 20 |
259756_at |
AT1G71080
|
[RNA polymerase II transcription elongation factor] |
0.586 |
-0.011 |
| 21 |
257714_at |
AT3G27360
|
H3.1, histone 3.1 |
0.586 |
0.089 |
| 22 |
262880_at |
AT1G64880
|
[Ribosomal protein S5 family protein] |
0.569 |
0.043 |
| 23 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
0.567 |
-0.208 |
| 24 |
245616_at |
AT4G14480
|
[Protein kinase superfamily protein] |
0.566 |
0.016 |
| 25 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
0.563 |
-0.126 |
| 26 |
259520_at |
AT1G12320
|
unknown |
0.561 |
-0.116 |
| 27 |
251791_at |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
0.561 |
0.012 |
| 28 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
0.558 |
-0.087 |
| 29 |
262480_at |
AT1G11340
|
[S-locus lectin protein kinase family protein] |
0.555 |
-0.034 |
| 30 |
260633_at |
AT1G62400
|
HT1, high leaf temperature 1 |
0.554 |
-0.060 |
| 31 |
256626_at |
AT3G20015
|
[Eukaryotic aspartyl protease family protein] |
0.553 |
-0.048 |
| 32 |
261745_at |
AT1G08500
|
AtENODL18, ENODL18, early nodulin-like protein 18 |
0.529 |
-0.035 |
| 33 |
260353_at |
AT1G69230
|
SP1L2, SPIRAL1-like2 |
0.528 |
0.049 |
| 34 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.524 |
0.256 |
| 35 |
250433_at |
AT5G10400
|
H3.1, histone 3.1 |
0.518 |
0.167 |
| 36 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
0.516 |
-0.002 |
| 37 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
0.511 |
0.078 |
| 38 |
262671_at |
AT1G76040
|
CPK29, calcium-dependent protein kinase 29 |
0.511 |
-0.045 |
| 39 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
0.496 |
-0.016 |
| 40 |
267628_at |
AT2G42280
|
AKS3, ABA-responsive kinase substrate 3, FBH4, FLOWERING BHLH 4 |
0.492 |
-0.052 |
| 41 |
260921_at |
AT1G21540
|
[AMP-dependent synthetase and ligase family protein] |
0.491 |
-0.004 |
| 42 |
261350_at |
AT1G79770
|
unknown |
0.484 |
-0.041 |
| 43 |
261146_at |
AT1G19620
|
unknown |
0.483 |
-0.055 |
| 44 |
257247_at |
AT3G24140
|
FMA, FAMA |
0.480 |
-0.098 |
| 45 |
247971_at |
AT5G56730
|
[Insulinase (Peptidase family M16) protein] |
0.480 |
0.037 |
| 46 |
256763_at |
AT3G16860
|
COBL8, COBRA-like protein 8 precursor |
0.480 |
0.074 |
| 47 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
0.477 |
-0.169 |
| 48 |
263047_at |
AT2G17630
|
PSAT2, phosphoserine aminotransferase 2 |
0.476 |
0.025 |
| 49 |
245692_at |
AT5G04150
|
BHLH101 |
0.470 |
0.233 |
| 50 |
262516_at |
AT1G17190
|
ATGSTU26, glutathione S-transferase tau 26, GSTU26, glutathione S-transferase tau 26 |
0.468 |
-0.143 |
| 51 |
249843_at |
AT5G23570
|
ATSGS3, SUPPRESSOR OF GENE SILENCING 3, SGS3, SUPPRESSOR OF GENE SILENCING 3 |
0.465 |
-0.030 |
| 52 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.462 |
-0.100 |
| 53 |
257421_at |
AT1G12030
|
unknown |
0.460 |
0.089 |
| 54 |
260546_at |
AT2G43520
|
ATTI2, trypsin inhibitor protein 2, TI2, trypsin inhibitor protein 2 |
0.460 |
0.040 |
| 55 |
263830_at |
AT2G40260
|
[Homeodomain-like superfamily protein] |
0.456 |
-0.100 |
| 56 |
252712_at |
AT3G43800
|
ATGSTU27, glutathione S-transferase tau 27, GSTU27, glutathione S-transferase tau 27 |
0.455 |
0.020 |
| 57 |
254663_at |
AT4G18290
|
KAT2, potassium channel in Arabidopsis thaliana 2 |
0.450 |
-0.053 |
| 58 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.448 |
-0.152 |
| 59 |
248369_at |
AT5G52040
|
At-RS41, arginine/serine-rich splicing factor 41, ATRSP41, RS41, arginine/serine-rich splicing factor 41 |
0.445 |
0.035 |
| 60 |
265002_at |
AT1G24400
|
AATL2, AMINO ACID TRANSPORTER-LIKE PROTEIN 2, ATLHT2, ARABIDOPSIS LYSINE HISTIDINE TRANSPORTER 2, LHT2, lysine histidine transporter 2 |
0.441 |
0.056 |
| 61 |
264947_at |
AT1G77020
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.437 |
-0.027 |
| 62 |
246860_at |
AT5G25840
|
unknown |
0.436 |
-0.039 |
| 63 |
262548_at |
AT1G31280
|
AGO2, argonaute 2, AtAGO2 |
0.436 |
0.218 |
| 64 |
257013_at |
AT3G26922
|
[F-box/RNI-like superfamily protein] |
0.435 |
-0.011 |
| 65 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
0.433 |
0.009 |
| 66 |
253162_at |
AT4G35630
|
PSAT1, phosphoserine aminotransferase 1 |
0.430 |
0.005 |
| 67 |
246861_at |
AT5G25890
|
IAA28, indole-3-acetic acid inducible 28, IAR2, IAA-ALANINE RESISTANT 2 |
0.426 |
-0.076 |
| 68 |
265596_at |
AT2G20020
|
ATCAF1, CAF1 |
0.425 |
0.018 |
| 69 |
266322_at |
AT2G46690
|
SAUR32, SMALL AUXIN UPREGULATED RNA 32 |
0.422 |
-0.169 |
| 70 |
248431_at |
AT5G51470
|
[Auxin-responsive GH3 family protein] |
0.419 |
-0.007 |
| 71 |
261059_at |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
0.413 |
-0.119 |
| 72 |
259514_at |
AT1G12480
|
CDI3, CARBON DIOXIDE INSENSITIVE 3, OZS1, OZONE-SENSITIVE 1, RCD3, RADICAL-INDUCED CELL DEATH 3, SLAC1, SLOW ANION CHANNEL-ASSOCIATED 1 |
0.413 |
0.057 |
| 73 |
248627_at |
AT5G48950
|
DHNAT2, DHNA-CoA thioesterase 2 |
0.412 |
0.010 |
| 74 |
251906_at |
AT3G53720
|
ATCHX20, cation/H+ exchanger 20, CHX20, cation/H+ exchanger 20 |
0.405 |
0.057 |
| 75 |
252858_at |
AT4G39770
|
TPPH, trehalose-6-phosphate phosphatase H |
0.405 |
-0.033 |
| 76 |
264525_at |
AT1G10060
|
ATBCAT-1, branched-chain amino acid transaminase 1, BCAT-1, branched-chain amino acid transaminase 1, BCAT1, branched-chain amino acid transferase 1 |
0.399 |
-0.027 |
| 77 |
244965_at |
ATCG00590
|
ORF31 |
-0.643 |
0.000 |
| 78 |
244966_at |
ATCG00600
|
PETG |
-0.414 |
-0.217 |
| 79 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
-0.375 |
-0.126 |
| 80 |
246273_at |
AT4G36700
|
[RmlC-like cupins superfamily protein] |
-0.348 |
0.573 |
| 81 |
266689_at |
AT2G19930
|
[RNA-dependent RNA polymerase family protein] |
-0.324 |
0.002 |
| 82 |
244967_at |
ATCG00630
|
PSAJ |
-0.303 |
-0.021 |
| 83 |
261818_at |
AT1G11390
|
[Protein kinase superfamily protein] |
-0.302 |
-0.005 |
| 84 |
258429_at |
AT3G16620
|
ATTOC120, ARABIDOPSIS THALIANA TRANSLOCON OUTER COMPLEX PROTEIN 120, TOC120, translocon outer complex protein 120 |
-0.285 |
0.010 |
| 85 |
249947_at |
AT5G19200
|
TSC10B, TSC10B |
-0.285 |
0.025 |
| 86 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
-0.278 |
-0.030 |
| 87 |
245025_at |
ATCG00130
|
ATPF |
-0.276 |
-0.050 |
| 88 |
260170_at |
AT1G71890
|
ATSUC5, SUCROSE-PROTON SYMPORTER 5, SUC5 |
-0.268 |
-0.070 |
| 89 |
249772_at |
AT5G24130
|
unknown |
-0.257 |
0.067 |
| 90 |
252911_at |
AT4G39510
|
CYP96A12, cytochrome P450, family 96, subfamily A, polypeptide 12 |
-0.256 |
0.027 |
| 91 |
244982_at |
ATCG00780
|
RPL14, ribosomal protein L14 |
-0.246 |
-0.084 |
| 92 |
250310_at |
AT5G12230
|
MED19A |
-0.245 |
0.019 |
| 93 |
262348_at |
AT2G48160
|
[Tudor/PWWP/MBT domain-containing protein] |
-0.243 |
-0.020 |
| 94 |
252854_at |
AT4G39370
|
UBP27, ubiquitin-specific protease 27 |
-0.238 |
-0.033 |
| 95 |
254174_at |
AT4G24120
|
ATYSL1, YELLOW STRIPE LIKE 1, YSL1, YELLOW STRIPE like 1 |
-0.236 |
-0.088 |
| 96 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
-0.235 |
-0.033 |
| 97 |
260472_at |
AT1G10990
|
unknown |
-0.232 |
-0.148 |
| 98 |
256325_at |
AT3G02330
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.231 |
0.098 |
| 99 |
262251_at |
AT1G53760
|
unknown |
-0.230 |
0.033 |
| 100 |
254185_at |
AT4G23990
|
ATCSLG3, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE G3, CSLG3, cellulose synthase like G3 |
-0.226 |
-0.058 |
| 101 |
253551_at |
AT4G30996
|
NKS1, NA(+)- AND K(+)-SENSITIVE 1 |
-0.224 |
0.016 |
| 102 |
265449_at |
AT2G46610
|
At-RS31a, arginine/serine-rich splicing factor 31a, RS31a, arginine/serine-rich splicing factor 31a |
-0.223 |
0.084 |
| 103 |
264907_at |
AT2G17280
|
[Phosphoglycerate mutase family protein] |
-0.221 |
0.059 |
| 104 |
262496_at |
AT1G21790
|
[TRAM, LAG1 and CLN8 (TLC) lipid-sensing domain containing protein] |
-0.220 |
-0.032 |
| 105 |
261597_at |
AT1G49780
|
PUB26, plant U-box 26 |
-0.220 |
-0.095 |
| 106 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
-0.215 |
0.054 |
| 107 |
245019_at |
ATCG00530
|
YCF10 |
-0.215 |
-0.038 |
| 108 |
254350_at |
AT4G22280
|
[F-box/RNI-like superfamily protein] |
-0.213 |
-0.018 |
| 109 |
261937_at |
AT1G22570
|
[Major facilitator superfamily protein] |
-0.212 |
0.007 |
| 110 |
264353_at |
AT1G03260
|
[SNARE associated Golgi protein family] |
-0.212 |
0.041 |
| 111 |
251630_at |
AT3G57420
|
unknown |
-0.210 |
-0.063 |
| 112 |
252189_at |
AT3G50070
|
CYCD3;3, CYCLIN D3;3 |
-0.208 |
0.023 |
| 113 |
251812_at |
AT3G54970
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
-0.201 |
0.101 |
| 114 |
245594_at |
AT4G14570
|
AARE, acylamino acid-releasing enzyme, AtAARE |
-0.199 |
0.010 |
| 115 |
254449_at |
AT4G20910
|
CRM2, CORYMBOSA 2, HEN1, HUA ENHANCER 1 |
-0.199 |
0.018 |
| 116 |
254333_at |
AT4G22753
|
ATSMO1-3, SMO1-3, sterol 4-alpha methyl oxidase 1-3 |
-0.198 |
0.064 |
| 117 |
265051_at |
AT1G52100
|
[Mannose-binding lectin superfamily protein] |
-0.198 |
-0.057 |
| 118 |
246586_at |
AT5G14770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.196 |
0.039 |
| 119 |
247703_at |
AT5G59560
|
SRR1, SENSITIVITY TO RED LIGHT REDUCED 1 |
-0.195 |
-0.031 |
| 120 |
258729_at |
AT3G11900
|
ANT1, aromatic and neutral transporter 1 |
-0.195 |
-0.104 |
| 121 |
266542_at |
AT2G35100
|
ARAD1, ARABINAN DEFICIENT 1 |
-0.195 |
-0.004 |
| 122 |
249452_at |
AT5G39500
|
ERMO1, ENDOPLASMIC RETICULUM MORPHOLOGY 1, GNL1, GNOM-like 1 |
-0.195 |
-0.046 |
| 123 |
245445_at |
AT4G16750
|
[Integrase-type DNA-binding superfamily protein] |
-0.193 |
0.001 |
| 124 |
260008_at |
AT1G68070
|
[Zinc finger, C3HC4 type (RING finger) family protein] |
-0.192 |
-0.017 |
| 125 |
253820_at |
AT4G28370
|
FLY1, FLYING SAUCER 1 |
-0.192 |
0.034 |
| 126 |
255885_at |
AT1G20330
|
CVP1, COTYLEDON VASCULAR PATTERN 1, FRL1, FRILL1, SMT2, sterol methyltransferase 2 |
-0.190 |
-0.045 |
| 127 |
257648_at |
AT3G16840
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.190 |
0.027 |
| 128 |
255349_at |
AT4G03830
|
unknown |
-0.188 |
0.015 |
| 129 |
266410_at |
AT2G38770
|
EMB2765, EMBRYO DEFECTIVE 2765 |
-0.188 |
0.021 |
| 130 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
-0.187 |
0.082 |
| 131 |
248269_at |
AT5G53470
|
ACBP, ACYL-COA BINDING PROTEIN, ACBP1, acyl-CoA binding protein 1, AtACBP1 |
-0.185 |
-0.012 |
| 132 |
264705_at |
AT1G09620
|
[ATP binding] |
-0.183 |
-0.014 |
| 133 |
261405_at |
AT1G18740
|
unknown |
-0.183 |
-0.087 |
| 134 |
255417_at |
AT4G03190
|
AFB1, AUXIN SIGNALING F BOX PROTEIN 1, ATGRH1, GRH1, GRR1-like protein 1 |
-0.183 |
0.009 |
| 135 |
244997_at |
ATCG00170
|
RPOC2 |
-0.182 |
-0.020 |
| 136 |
262338_at |
AT1G64185
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
-0.182 |
0.026 |
| 137 |
255638_at |
AT4G00740
|
QUA3, QUASIMODO 3 |
-0.182 |
-0.025 |
| 138 |
257616_at |
AT3G26540
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.182 |
0.047 |
| 139 |
245558_at |
AT4G15430
|
[ERD (early-responsive to dehydration stress) family protein] |
-0.181 |
-0.112 |
| 140 |
261377_at |
AT1G18850
|
unknown |
-0.179 |
0.024 |
| 141 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
-0.179 |
0.022 |
| 142 |
245017_at |
ATCG00510
|
PSAI, photsystem I subunit I |
-0.178 |
-0.062 |
| 143 |
266774_at |
AT2G29050
|
ATRBL1, RHOMBOID-like 1, RBL1, RHOMBOID-like 1 |
-0.177 |
0.007 |
| 144 |
264116_at |
AT2G31320
|
PARP1, POLY(ADP-RIBOSE) POLYMERASE 1 |
-0.176 |
0.051 |
| 145 |
265663_at |
AT2G24290
|
unknown |
-0.176 |
-0.059 |
| 146 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
-0.175 |
0.007 |
| 147 |
253887_at |
AT4G27730
|
ATOPT6, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 6, OPT6, oligopeptide transporter 1 |
-0.174 |
-0.031 |
| 148 |
250972_at |
AT5G02840
|
LCL1, LHY/CCA1-like 1 |
-0.173 |
-0.043 |
| 149 |
265424_at |
AT2G20780
|
[Major facilitator superfamily protein] |
-0.173 |
-0.042 |
| 150 |
258323_at |
AT3G22750
|
[Protein kinase superfamily protein] |
-0.171 |
0.014 |
| 151 |
255036_at |
AT4G09560
|
[Protease-associated (PA) RING/U-box zinc finger family protein] |
-0.171 |
-0.018 |