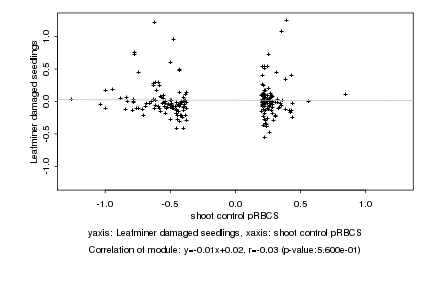
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
shoot control pRBCS |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
245385_at |
AT4G14020
|
[Rapid alkalinization factor (RALF) family protein] |
0.838 |
0.114 |
| 2 |
260672_at |
AT1G19480
|
[DNA glycosylase superfamily protein] |
0.557 |
0.001 |
| 3 |
258315_at |
AT3G16175
|
[Thioesterase superfamily protein] |
0.436 |
-0.237 |
| 4 |
247024_at |
AT5G66985
|
unknown |
0.435 |
-0.016 |
| 5 |
262902_x_at |
AT1G59930
|
[MADS-box family protein] |
0.429 |
-0.132 |
| 6 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.426 |
0.402 |
| 7 |
247291_at |
AT5G64480
|
unknown |
0.421 |
-0.155 |
| 8 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
0.412 |
-0.129 |
| 9 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.387 |
1.263 |
| 10 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
0.379 |
0.353 |
| 11 |
246518_at |
AT5G15770
|
AtGNA1, glucose-6-phosphate acetyltransferase 1, GNA1, glucose-6-phosphate acetyltransferase 1 |
0.377 |
-0.118 |
| 12 |
259423_at |
AT1G13880
|
[ELM2 domain-containing protein] |
0.358 |
0.029 |
| 13 |
260076_at |
AT1G73630
|
[EF hand calcium-binding protein family] |
0.351 |
-0.046 |
| 14 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.347 |
1.092 |
| 15 |
252661_at |
AT3G44450
|
unknown |
0.339 |
-0.018 |
| 16 |
250650_at |
AT5G06850
|
FTIP1, FT-interacting protein 1 |
0.335 |
-0.083 |
| 17 |
264974_at |
AT1G27050
|
[homeobox protein 54 (HB54)] |
0.324 |
-0.101 |
| 18 |
246653_at |
AT5G35200
|
[ENTH/ANTH/VHS superfamily protein] |
0.316 |
0.038 |
| 19 |
251985_at |
AT3G53220
|
[Thioredoxin superfamily protein] |
0.316 |
-0.013 |
| 20 |
261938_at |
AT1G22510
|
[FUNCTIONS IN: zinc ion binding] |
0.312 |
0.454 |
| 21 |
247656_at |
AT5G59890
|
ADF4, actin depolymerizing factor 4, ATADF4 |
0.306 |
-0.002 |
| 22 |
245747_at |
AT1G51100
|
CRR41, CHLORORESPIRATORY REDUCTION 41 |
0.302 |
-0.226 |
| 23 |
253940_at |
AT4G26950
|
unknown |
0.301 |
-0.205 |
| 24 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
0.291 |
-0.278 |
| 25 |
258928_at |
AT3G10070
|
TAF12, TBP-associated factor 12, TAFII58, TATA-ASSOCIATED FACTOR II 58 |
0.283 |
0.005 |
| 26 |
261286_at |
AT1G35780
|
unknown |
0.282 |
-0.091 |
| 27 |
247255_at |
AT5G64780
|
[Uncharacterised conserved protein UCP009193] |
0.280 |
-0.032 |
| 28 |
247836_at |
AT5G57860
|
[Ubiquitin-like superfamily protein] |
0.280 |
0.090 |
| 29 |
263593_at |
AT2G01860
|
EMB975, EMBRYO DEFECTIVE 975 |
0.280 |
-0.181 |
| 30 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
0.274 |
0.099 |
| 31 |
246007_at |
AT5G08410
|
FTRA2, ferredoxin/thioredoxin reductase subunit A (variable subunit) 2 |
0.272 |
-0.005 |
| 32 |
247632_at |
AT5G60460
|
[Preprotein translocase Sec, Sec61-beta subunit protein] |
0.271 |
0.070 |
| 33 |
251454_at |
AT3G60080
|
[RING/U-box superfamily protein] |
0.270 |
-0.131 |
| 34 |
254661_at |
AT4G18260
|
[Cytochrome b561/ferric reductase transmembrane protein family] |
0.269 |
0.070 |
| 35 |
250603_at |
AT5G07820
|
[Plant calmodulin-binding protein-related] |
0.267 |
0.031 |
| 36 |
265044_at |
AT1G03950
|
VPS2.3, vacuolar protein sorting-associated protein 2.3 |
0.266 |
0.135 |
| 37 |
245337_at |
AT4G16566
|
HINT4, histidine triad nucleotide-binding 4 |
0.265 |
-0.023 |
| 38 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
0.258 |
-0.467 |
| 39 |
249404_at |
AT5G40160
|
EMB139, EMBRYO DEFECTIVE 139, EMB506, embryo defective 506 |
0.256 |
-0.055 |
| 40 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
0.256 |
-0.065 |
| 41 |
259497_at |
AT1G15860
|
unknown |
0.254 |
0.057 |
| 42 |
265226_at |
AT2G28430
|
unknown |
0.254 |
0.010 |
| 43 |
254276_at |
AT4G22820
|
[A20/AN1-like zinc finger family protein] |
0.250 |
0.205 |
| 44 |
267070_at |
AT2G41000
|
[Chaperone DnaJ-domain superfamily protein] |
0.248 |
-0.108 |
| 45 |
245545_at |
AT4G15280
|
UGT71B5, UDP-glucosyl transferase 71B5 |
0.247 |
0.740 |
| 46 |
251727_at |
AT3G56290
|
unknown |
0.244 |
-0.250 |
| 47 |
250515_at |
AT5G09570
|
[Cox19-like CHCH family protein] |
0.244 |
-0.027 |
| 48 |
260367_at |
AT1G69760
|
unknown |
0.242 |
0.032 |
| 49 |
261733_at |
AT1G47830
|
[SNARE-like superfamily protein] |
0.240 |
0.106 |
| 50 |
256767_at |
AT3G13680
|
[F-box and associated interaction domains-containing protein] |
0.239 |
-0.019 |
| 51 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
0.239 |
0.545 |
| 52 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
0.239 |
-0.097 |
| 53 |
261516_at |
AT1G71750
|
HGPT, Hypoxanthine-guanine phosphoribosyltransferase |
0.239 |
0.024 |
| 54 |
246159_at |
AT5G20935
|
CRR42, CHLORORESPIRATORY REDUCTION 42 |
0.238 |
-0.379 |
| 55 |
260427_at |
AT1G72430
|
SAUR78, SMALL AUXIN UPREGULATED RNA 78 |
0.234 |
-0.377 |
| 56 |
262196_at |
AT1G77870
|
MUB5, membrane-anchored ubiquitin-fold protein 5 precursor |
0.233 |
-0.349 |
| 57 |
257698_at |
AT3G12730
|
[Homeodomain-like superfamily protein] |
0.231 |
0.018 |
| 58 |
259659_at |
AT1G55170
|
unknown |
0.230 |
0.065 |
| 59 |
247754_at |
AT5G59080
|
unknown |
0.228 |
0.178 |
| 60 |
258920_at |
AT3G10520
|
AHB2, haemoglobin 2, ARATH GLB2, ATGLB2, ARABIDOPSIS HEMOGLOBIN 2, GLB2, HEMOGLOBIN 2, HB2, haemoglobin 2, NSHB2, NON-SYMBIOTIC HAEMOGLOBIN 2 |
0.227 |
-0.337 |
| 61 |
260622_at |
AT1G07980
|
NF-YC10, nuclear factor Y, subunit C10 |
0.227 |
-0.023 |
| 62 |
262160_at |
AT1G52590
|
[Putative thiol-disulphide oxidoreductase DCC] |
0.227 |
-0.126 |
| 63 |
250353_at |
AT5G11630
|
NOXY2, nonresponding to oxylipins 2 |
0.226 |
-0.014 |
| 64 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.225 |
-0.292 |
| 65 |
247420_at |
AT5G63100
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.223 |
-0.273 |
| 66 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
0.222 |
0.518 |
| 67 |
252063_at |
AT3G51590
|
LTP12, lipid transfer protein 12 |
0.221 |
0.122 |
| 68 |
265214_at |
AT1G05000
|
AtPFA-DSP1, PFA-DSP1, plant and fungi atypical dual-specificity phosphatase 1 |
0.220 |
0.178 |
| 69 |
263331_at |
AT2G15270
|
unknown |
0.220 |
0.074 |
| 70 |
267467_at |
AT2G30580
|
BMI1A, DRIP2, DREB2A-interacting protein 2 |
0.218 |
-0.074 |
| 71 |
265183_at |
AT1G23750
|
[Nucleic acid-binding, OB-fold-like protein] |
0.218 |
-0.033 |
| 72 |
252565_at |
AT3G46000
|
ADF2, actin depolymerizing factor 2 |
0.217 |
0.123 |
| 73 |
249087_at |
AT5G44210
|
ATERF-9, ERF DOMAIN PROTEIN- 9, ATERF9, ERF DOMAIN PROTEIN 9, ERF9, erf domain protein 9 |
0.217 |
0.543 |
| 74 |
249878_at |
AT5G23090
|
NF-YB13, nuclear factor Y, subunit B13 |
0.216 |
0.058 |
| 75 |
251008_at |
AT5G02710
|
unknown |
0.216 |
-0.174 |
| 76 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
0.216 |
-0.545 |
| 77 |
246218_at |
AT4G36800
|
RCE1, RUB1 conjugating enzyme 1 |
0.215 |
0.013 |
| 78 |
258640_at |
AT3G07930
|
[DNA glycosylase superfamily protein] |
0.214 |
-0.047 |
| 79 |
246140_at |
AT5G19910
|
MED31 |
0.214 |
-0.031 |
| 80 |
262974_at |
AT1G75550
|
[glycine-rich protein] |
0.213 |
-0.025 |
| 81 |
253302_at |
AT4G33660
|
unknown |
0.212 |
-0.221 |
| 82 |
247753_at |
AT5G59070
|
[UDP-Glycosyltransferase superfamily protein] |
0.211 |
0.124 |
| 83 |
258915_at |
AT3G10640
|
VPS60.1 |
0.211 |
0.248 |
| 84 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
0.210 |
-0.359 |
| 85 |
252373_at |
AT3G48090
|
ATEDS1, EDS1, enhanced disease susceptibility 1 |
0.210 |
0.028 |
| 86 |
254611_at |
AT4G19095
|
unknown |
0.210 |
-0.046 |
| 87 |
248379_at |
AT5G51700
|
ATRAR1, PBS2, PPHB SUSCEPTIBLE 2, RAR1, Required for Mla12 resistance 1, RPR2 |
0.210 |
0.084 |
| 88 |
248632_at |
AT5G49010
|
EMB2812, EMBRYO DEFECTIVE 2812, SLD5, SYNTHETIC LETHALITY WITH DPB11-1 5 |
0.209 |
0.083 |
| 89 |
252223_at |
AT3G49850
|
ATTRB3, TBP2, TELOMERE-BINDING PROTEIN 2, TRB3, telomere repeat binding factor 3 |
0.209 |
-0.112 |
| 90 |
265510_at |
AT2G05630
|
ATG8D |
0.208 |
0.030 |
| 91 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.208 |
0.105 |
| 92 |
251986_at |
AT3G53310
|
[AP2/B3-like transcriptional factor family protein] |
0.207 |
0.122 |
| 93 |
265093_at |
AT1G03905
|
ABCI19, ATP-binding cassette I19 |
0.207 |
0.272 |
| 94 |
254891_at |
AT4G11740
|
SAY1 |
0.207 |
-0.036 |
| 95 |
262107_at |
AT1G02750
|
[Drought-responsive family protein] |
0.207 |
-0.072 |
| 96 |
252650_at |
AT3G44690
|
unknown |
0.206 |
0.020 |
| 97 |
256384_at |
AT1G66660
|
[Protein with RING/U-box and TRAF-like domains] |
0.205 |
0.001 |
| 98 |
258972_at |
AT3G01920
|
[DHBP synthase RibB-like alpha/beta domain] |
0.205 |
-0.050 |
| 99 |
261487_at |
AT1G14340
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.204 |
-0.002 |
| 100 |
265913_at |
AT2G25625
|
unknown |
0.204 |
0.111 |
| 101 |
262622_at |
AT1G06510
|
unknown |
0.204 |
-0.012 |
| 102 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
0.204 |
0.540 |
| 103 |
251197_at |
AT3G62960
|
[Thioredoxin superfamily protein] |
0.203 |
0.043 |
| 104 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
0.202 |
0.414 |
| 105 |
267123_at |
AT2G23560
|
ATMES7, ARABIDOPSIS THALIANA METHYL ESTERASE 7, MES7, methyl esterase 7 |
0.200 |
0.102 |
| 106 |
266570_at |
AT2G24090
|
PRPL35, plastid ribosomal protein L35 |
0.198 |
-0.141 |
| 107 |
266393_at |
AT2G41260
|
ATM17, M17 |
-1.264 |
0.037 |
| 108 |
266353_at |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
-1.038 |
-0.036 |
| 109 |
265211_at |
AT2G36640
|
ATECP63, embryonic cell protein 63, ECP63, embryonic cell protein 63 |
-1.003 |
0.170 |
| 110 |
244981_at |
ATCG00770
|
RPS8, ribosomal protein S8 |
-0.999 |
-0.095 |
| 111 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
-0.946 |
0.189 |
| 112 |
256814_at |
AT3G21370
|
BGLU19, beta glucosidase 19 |
-0.886 |
0.048 |
| 113 |
244985_at |
ATCG00810
|
RPL22, ribosomal protein L22 |
-0.848 |
-0.110 |
| 114 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.841 |
0.075 |
| 115 |
252019_at |
AT3G53040
|
[late embryogenesis abundant protein, putative / LEA protein, putative] |
-0.832 |
0.014 |
| 116 |
245049_at |
ATCG00050
|
RPS16, ribosomal protein S16 |
-0.794 |
-0.133 |
| 117 |
251838_at |
AT3G54940
|
[Papain family cysteine protease] |
-0.788 |
0.036 |
| 118 |
250624_at |
AT5G07330
|
unknown |
-0.784 |
0.011 |
| 119 |
244939_at |
ATCG00065
|
RPS12, RIBOSOMAL PROTEIN S12, RPS12A, ribosomal protein S12A |
-0.783 |
-0.007 |
| 120 |
256464_at |
AT1G32560
|
AtLEA4-1, Late Embryogenesis Abundant 4-1, LEA4-1, Late Embryogenesis Abundant 4-1 |
-0.779 |
0.758 |
| 121 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
-0.778 |
0.729 |
| 122 |
244980_at |
ATCG00760
|
RPL36, ribosomal protein L36 |
-0.767 |
-0.104 |
| 123 |
244966_at |
ATCG00600
|
PETG |
-0.751 |
-0.107 |
| 124 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
-0.750 |
0.456 |
| 125 |
245016_at |
ATCG00500
|
ACCD, acetyl-CoA carboxylase carboxyl transferase subunit beta |
-0.717 |
-0.109 |
| 126 |
245008_at |
ATCG00360
|
YCF3 |
-0.709 |
-0.213 |
| 127 |
244932_at |
ATCG01060
|
PSAC |
-0.692 |
-0.063 |
| 128 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
-0.688 |
-0.016 |
| 129 |
256354_at |
AT1G54870
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.663 |
-0.016 |
| 130 |
244970_at |
ATCG00660
|
RPL20, ribosomal protein L20 |
-0.661 |
-0.026 |
| 131 |
245555_at |
AT4G15390
|
[HXXXD-type acyl-transferase family protein] |
-0.646 |
0.009 |
| 132 |
247437_at |
AT5G62490
|
ATHVA22B, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE B, HVA22B, HVA22 homologue B |
-0.636 |
0.046 |
| 133 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
-0.633 |
0.288 |
| 134 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
-0.630 |
0.261 |
| 135 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
-0.628 |
1.222 |
| 136 |
245011_at |
ATCG00430
|
PSBG, photosystem II reaction center protein G |
-0.623 |
-0.096 |
| 137 |
250868_at |
AT5G03860
|
MLS, malate synthase |
-0.619 |
0.016 |
| 138 |
244933_at |
ATCG01070
|
NDHE |
-0.619 |
-0.051 |
| 139 |
253392_at |
AT4G32650
|
ATKC1, ARABIDOPSIS THALIANA K+ RECTIFYING CHANNEL 1, AtLKT1, A. thaliana low-K+-tolerant 1, KAT3, potassium channel in Arabidopsis thaliana 3, KC1 |
-0.616 |
0.294 |
| 140 |
250500_at |
AT5G09530
|
PELPK1, Pro-Glu-Leu|Ile|Val-Pro-Lys 1, PRP10, proline-rich protein 10 |
-0.608 |
0.178 |
| 141 |
256338_at |
AT1G72100
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
-0.597 |
-0.069 |
| 142 |
251942_at |
AT3G53480
|
ABCG37, ATP-binding cassette G37, ATPDR9, PLEIOTROPIC DRUG RESISTANCE 9, PDR9, pleiotropic drug resistance 9, PIS1, polar auxin transport inhibitor sensitive 1 |
-0.594 |
0.299 |
| 143 |
245017_at |
ATCG00510
|
PSAI, photsystem I subunit I |
-0.587 |
-0.102 |
| 144 |
247297_at |
AT5G64100
|
[Peroxidase superfamily protein] |
-0.586 |
0.247 |
| 145 |
244982_at |
ATCG00780
|
RPL14, ribosomal protein L14 |
-0.580 |
-0.153 |
| 146 |
257334_at |
ATMG01370
|
ORF111D |
-0.576 |
0.079 |
| 147 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
-0.573 |
0.068 |
| 148 |
245021_at |
ATCG00550
|
PSBJ, photosystem II reaction center protein J |
-0.561 |
-0.016 |
| 149 |
244963_at |
ATCG00570
|
PSBF, photosystem II reaction center protein F |
-0.558 |
-0.014 |
| 150 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
-0.556 |
0.057 |
| 151 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
-0.553 |
0.094 |
| 152 |
244984_at |
ATCG00800
|
[RESISTANCE TO PSEUDOMONAS SYRINGAE 3 (RPS3)] |
-0.549 |
-0.069 |
| 153 |
260088_at |
AT1G73190
|
ALPHA-TIP, ALPHA-TONOPLAST INTRINSIC PROTEIN, TIP3;1 |
-0.543 |
-0.017 |
| 154 |
253191_at |
AT4G35350
|
XCP1, xylem cysteine peptidase 1 |
-0.542 |
-0.175 |
| 155 |
248728_at |
AT5G48000
|
CYP708 A2, CYTOCHROME P450, FAMILY 708, SUBFAMILY A, POLYPEPTIDE 2, CYP708A2, cytochrome P450, family 708, subfamily A, polypeptide 2, THAH, THALIANOL HYDROXYLASE, THAH1, THALIANOL HYDROXYLASE 1 |
-0.541 |
-0.025 |
| 156 |
266143_at |
AT2G38905
|
[Low temperature and salt responsive protein family] |
-0.540 |
0.005 |
| 157 |
244996_at |
ATCG00160
|
RPS2, ribosomal protein S2 |
-0.530 |
-0.104 |
| 158 |
244934_at |
ATCG01080
|
NDHG |
-0.526 |
-0.080 |
| 159 |
244964_at |
ATCG00580
|
PSBE, photosystem II reaction center protein E |
-0.516 |
-0.032 |
| 160 |
248727_at |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
-0.503 |
0.026 |
| 161 |
248912_at |
AT5G45670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.501 |
-0.270 |
| 162 |
244979_at |
ATCG00750
|
RPS11, ribosomal protein S11 |
-0.500 |
-0.057 |
| 163 |
245010_at |
ATCG00420
|
NDHJ, NADH dehydrogenase subunit J |
-0.500 |
-0.085 |
| 164 |
249045_at |
AT5G44380
|
[FAD-binding Berberine family protein] |
-0.499 |
0.612 |
| 165 |
263098_at |
AT2G16005
|
[MD-2-related lipid recognition domain-containing protein] |
-0.496 |
0.027 |
| 166 |
266941_at |
AT2G18980
|
[Peroxidase superfamily protein] |
-0.495 |
-0.020 |
| 167 |
264166_at |
AT1G65370
|
[TRAF-like family protein] |
-0.490 |
-0.099 |
| 168 |
260166_at |
AT1G79840
|
GL2, GLABRA 2 |
-0.486 |
-0.068 |
| 169 |
244936_at |
ATCG01100
|
NDHA |
-0.485 |
-0.072 |
| 170 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
-0.482 |
0.967 |
| 171 |
244998_at |
ATCG00180
|
RPOC1 |
-0.480 |
-0.075 |
| 172 |
245018_at |
ATCG00520
|
YCF4 |
-0.476 |
-0.101 |
| 173 |
245019_at |
ATCG00530
|
YCF10 |
-0.462 |
-0.066 |
| 174 |
245015_at |
ATCG00490
|
RBCL |
-0.460 |
-0.117 |
| 175 |
244997_at |
ATCG00170
|
RPOC2 |
-0.458 |
-0.174 |
| 176 |
247061_at |
AT5G66780
|
unknown |
-0.457 |
-0.007 |
| 177 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
-0.455 |
-0.138 |
| 178 |
258863_at |
AT3G03300
|
ATDCL2, DICER-LIKE 2, DCL2, dicer-like 2 |
-0.455 |
-0.180 |
| 179 |
254633_at |
AT4G18640
|
MRH1, morphogenesis of root hair 1 |
-0.454 |
-0.403 |
| 180 |
263485_at |
AT2G29890
|
ATVLN1, VLN1, villin 1 |
-0.449 |
-0.266 |
| 181 |
261845_at |
AT1G15960
|
ATNRAMP6, NRAMP6, NRAMP metal ion transporter 6 |
-0.447 |
-0.211 |
| 182 |
267645_at |
AT2G32860
|
BGLU33, beta glucosidase 33 |
-0.444 |
-0.139 |
| 183 |
245022_at |
ATCG00560
|
PSBL, photosystem II reaction center protein L |
-0.441 |
-0.043 |
| 184 |
245009_at |
ATCG00380
|
RPS4, chloroplast ribosomal protein S4 |
-0.441 |
-0.031 |
| 185 |
260890_at |
AT1G29090
|
[Cysteine proteinases superfamily protein] |
-0.438 |
-0.046 |
| 186 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
-0.436 |
0.140 |
| 187 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
-0.435 |
0.487 |
| 188 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
-0.433 |
0.494 |
| 189 |
244986_at |
ATCG00820
|
RPS19, ribosomal protein S19 |
-0.433 |
-0.064 |
| 190 |
261653_at |
AT1G01900
|
ATSBT1.1, SBTI1.1 |
-0.431 |
-0.298 |
| 191 |
261930_at |
AT1G22440
|
[Zinc-binding alcohol dehydrogenase family protein] |
-0.427 |
-0.037 |
| 192 |
252537_at |
AT3G45710
|
[Major facilitator superfamily protein] |
-0.427 |
-0.018 |
| 193 |
249542_at |
AT5G38140
|
NF-YC12, nuclear factor Y, subunit C12 |
-0.422 |
-0.203 |
| 194 |
259615_at |
AT1G47980
|
unknown |
-0.420 |
0.012 |
| 195 |
257217_at |
AT3G14940
|
ATPPC3, phosphoenolpyruvate carboxylase 3, PPC3, phosphoenolpyruvate carboxylase 3 |
-0.420 |
-0.212 |
| 196 |
254327_at |
AT4G22490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.415 |
-0.003 |
| 197 |
244937_at |
ATCG01110
|
NDHH, NAD(P)H dehydrogenase subunit H |
-0.411 |
-0.243 |
| 198 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.402 |
0.004 |
| 199 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
-0.402 |
0.062 |
| 200 |
262796_at |
AT1G20850
|
XCP2, xylem cysteine peptidase 2 |
-0.401 |
-0.134 |
| 201 |
264469_at |
AT1G67100
|
LBD40, LOB domain-containing protein 40 |
-0.401 |
-0.077 |
| 202 |
265250_at |
AT2G01950
|
BRL2, BRI1-like 2, VH1, VASCULAR HIGHWAY 1 |
-0.400 |
-0.409 |
| 203 |
256321_at |
AT1G55020
|
ATLOX1, ARABIDOPSIS LIPOXYGENASE 1, LOX1, lipoxygenase 1 |
-0.395 |
0.029 |
| 204 |
245014_at |
ATCG00480
|
ATPB, ATP synthase subunit beta, PB, ATP synthase subunit beta |
-0.394 |
-0.031 |
| 205 |
250228_at |
AT5G13840
|
CCS52B, cell cycle switch protein 52 B, FZR3, FIZZY-related 3 |
-0.392 |
-0.066 |
| 206 |
261991_at |
AT1G33700
|
[Beta-glucosidase, GBA2 type family protein] |
-0.388 |
-0.211 |
| 207 |
253753_at |
AT4G29030
|
[Putative membrane lipoprotein] |
-0.387 |
0.122 |
| 208 |
248729_at |
AT5G48010
|
AtTHAS1, Arabidopsis thaliana thalianol synthase 1, THAS, THALIANOL SYNTHASE, THAS1, thalianol synthase 1 |
-0.385 |
0.020 |
| 209 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.383 |
-0.284 |
| 210 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.382 |
0.150 |
| 211 |
244962_at |
ATCG01050
|
NDHD |
-0.381 |
-0.093 |
| 212 |
265902_at |
AT2G25590
|
[Plant Tudor-like protein] |
-0.376 |
-0.009 |