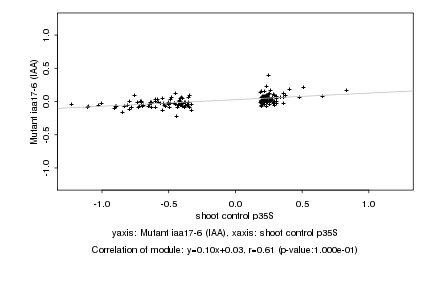
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
shoot control p35S |
Log2 signal ratio
Mutant iaa17-6 (IAA) |
| 1 |
245385_at |
AT4G14020
|
[Rapid alkalinization factor (RALF) family protein] |
0.827 |
0.176 |
| 2 |
258201_at |
AT3G13910
|
unknown |
0.649 |
0.090 |
| 3 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
0.509 |
0.218 |
| 4 |
260025_at |
AT1G30070
|
[SGS domain-containing protein] |
0.476 |
0.061 |
| 5 |
255028_at |
AT4G09890
|
unknown |
0.401 |
0.184 |
| 6 |
246270_at |
AT4G36500
|
unknown |
0.370 |
0.103 |
| 7 |
246518_at |
AT5G15770
|
AtGNA1, glucose-6-phosphate acetyltransferase 1, GNA1, glucose-6-phosphate acetyltransferase 1 |
0.357 |
0.075 |
| 8 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.357 |
0.123 |
| 9 |
260076_at |
AT1G73630
|
[EF hand calcium-binding protein family] |
0.354 |
-0.029 |
| 10 |
261259_at |
AT1G26660
|
[Prefoldin chaperone subunit family protein] |
0.331 |
0.065 |
| 11 |
258617_at |
AT3G03000
|
[EF hand calcium-binding protein family] |
0.307 |
-0.016 |
| 12 |
260672_at |
AT1G19480
|
[DNA glycosylase superfamily protein] |
0.303 |
0.004 |
| 13 |
253726_at |
AT4G29430
|
rps15ae, ribosomal protein S15A E |
0.303 |
0.051 |
| 14 |
247836_at |
AT5G57860
|
[Ubiquitin-like superfamily protein] |
0.303 |
0.085 |
| 15 |
247255_at |
AT5G64780
|
[Uncharacterised conserved protein UCP009193] |
0.291 |
0.096 |
| 16 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
0.290 |
-0.054 |
| 17 |
250353_at |
AT5G11630
|
NOXY2, nonresponding to oxylipins 2 |
0.287 |
0.005 |
| 18 |
264974_at |
AT1G27050
|
[homeobox protein 54 (HB54)] |
0.283 |
-0.016 |
| 19 |
262838_at |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.281 |
-0.006 |
| 20 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
0.280 |
0.116 |
| 21 |
262286_at |
AT1G68585
|
unknown |
0.275 |
0.072 |
| 22 |
266326_at |
AT2G46650
|
ATCB5-C, ARABIDOPSIS CYTOCHROME B5 ISOFORM C, B5 #1, CB5-C, cytochrome B5 isoform C, CYTB5-C |
0.270 |
0.027 |
| 23 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
0.269 |
0.036 |
| 24 |
252352_at |
AT3G48185
|
unknown |
0.268 |
0.040 |
| 25 |
251197_at |
AT3G62960
|
[Thioredoxin superfamily protein] |
0.261 |
0.013 |
| 26 |
260040_at |
AT1G68765
|
IDA, INFLORESCENCE DEFICIENT IN ABSCISSION |
0.259 |
0.179 |
| 27 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
0.257 |
0.045 |
| 28 |
262196_at |
AT1G77870
|
MUB5, membrane-anchored ubiquitin-fold protein 5 precursor |
0.256 |
0.023 |
| 29 |
254362_at |
AT4G22160
|
unknown |
0.255 |
0.051 |
| 30 |
256720_at |
AT2G34140
|
[Dof-type zinc finger DNA-binding family protein] |
0.252 |
0.122 |
| 31 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
0.248 |
-0.031 |
| 32 |
248632_at |
AT5G49010
|
EMB2812, EMBRYO DEFECTIVE 2812, SLD5, SYNTHETIC LETHALITY WITH DPB11-1 5 |
0.247 |
0.009 |
| 33 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
0.247 |
0.106 |
| 34 |
253044_at |
AT4G37290
|
unknown |
0.242 |
0.395 |
| 35 |
246429_at |
AT5G17450
|
HIPP21, heavy metal associated isoprenylated plant protein 21 |
0.239 |
0.075 |
| 36 |
249087_at |
AT5G44210
|
ATERF-9, ERF DOMAIN PROTEIN- 9, ATERF9, ERF DOMAIN PROTEIN 9, ERF9, erf domain protein 9 |
0.239 |
0.104 |
| 37 |
265937_at |
AT2G19610
|
[RING/U-box superfamily protein] |
0.238 |
-0.003 |
| 38 |
265226_at |
AT2G28430
|
unknown |
0.237 |
0.025 |
| 39 |
254201_at |
AT4G24130
|
unknown |
0.236 |
0.095 |
| 40 |
247146_at |
AT5G65610
|
unknown |
0.236 |
0.087 |
| 41 |
257081_at |
AT3G30460
|
[RING/U-box superfamily protein] |
0.233 |
0.017 |
| 42 |
249822_at |
AT5G23710
|
[DNA binding] |
0.230 |
0.026 |
| 43 |
254761_at |
AT4G13195
|
CLE44, CLAVATA3/ESR-RELATED 44 |
0.230 |
0.228 |
| 44 |
260622_at |
AT1G07980
|
NF-YC10, nuclear factor Y, subunit C10 |
0.229 |
-0.000 |
| 45 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
0.228 |
0.095 |
| 46 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
0.227 |
-0.063 |
| 47 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.225 |
0.084 |
| 48 |
260367_at |
AT1G69760
|
unknown |
0.221 |
0.085 |
| 49 |
259095_at |
AT3G05020
|
ACP, ACYL CARRIER PROTEIN, ACP1, acyl carrier protein 1 |
0.220 |
0.016 |
| 50 |
245337_at |
AT4G16566
|
HINT4, histidine triad nucleotide-binding 4 |
0.220 |
0.018 |
| 51 |
259822_at |
AT1G66230
|
AtMYB20, myb domain protein 20, MYB20, myb domain protein 20 |
0.219 |
0.041 |
| 52 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
0.219 |
0.066 |
| 53 |
253165_at |
AT4G35320
|
unknown |
0.217 |
-0.030 |
| 54 |
262828_at |
AT1G14950
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.214 |
0.019 |
| 55 |
257517_at |
AT3G16330
|
unknown |
0.213 |
0.007 |
| 56 |
263693_at |
AT1G31200
|
ATPP2-A9, phloem protein 2-A9, PP2-A9, phloem protein 2-A9 |
0.213 |
0.050 |
| 57 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
0.213 |
0.089 |
| 58 |
252224_at |
AT3G49860
|
ARLA1B, ADP-ribosylation factor-like A1B, ATARLA1B, ADP-ribosylation factor-like A1B |
0.212 |
0.165 |
| 59 |
250740_at |
AT5G05760
|
ATSED5, ATSYP31, SED5, T-SNARE SED 5, SYP31, syntaxin of plants 31 |
0.207 |
0.078 |
| 60 |
261409_at |
AT1G07640
|
OBP2, URP3, UAS-TAGGED ROOT PATTERNING3 |
0.207 |
0.055 |
| 61 |
248069_at |
AT5G55650
|
unknown |
0.207 |
-0.000 |
| 62 |
265732_at |
AT2G01300
|
unknown |
0.205 |
0.017 |
| 63 |
265510_at |
AT2G05630
|
ATG8D |
0.205 |
0.047 |
| 64 |
256880_at |
AT3G26450
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.202 |
0.036 |
| 65 |
250722_at |
AT5G06190
|
unknown |
0.201 |
0.077 |
| 66 |
246054_at |
AT5G08360
|
unknown |
0.201 |
0.029 |
| 67 |
250734_at |
AT5G06270
|
unknown |
0.198 |
-0.044 |
| 68 |
252565_at |
AT3G46000
|
ADF2, actin depolymerizing factor 2 |
0.198 |
0.023 |
| 69 |
246581_at |
AT1G31760
|
[SWIB/MDM2 domain superfamily protein] |
0.195 |
0.012 |
| 70 |
248512_at |
AT5G50460
|
[secE/sec61-gamma protein transport protein] |
0.193 |
0.022 |
| 71 |
252222_at |
AT3G49845
|
WIH3, WINDHOSE 3 |
0.190 |
0.006 |
| 72 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
0.190 |
0.158 |
| 73 |
254656_at |
AT4G18070
|
unknown |
0.189 |
-0.017 |
| 74 |
245445_at |
AT4G16750
|
[Integrase-type DNA-binding superfamily protein] |
0.189 |
-0.054 |
| 75 |
250158_at |
AT5G15190
|
unknown |
0.189 |
0.004 |
| 76 |
248646_at |
AT5G49100
|
unknown |
0.188 |
0.070 |
| 77 |
253629_at |
AT4G30450
|
[glycine-rich protein] |
0.188 |
-0.060 |
| 78 |
267463_at |
AT2G33845
|
[Nucleic acid-binding, OB-fold-like protein] |
0.188 |
0.022 |
| 79 |
262438_at |
AT1G47410
|
unknown |
0.187 |
0.143 |
| 80 |
256145_at |
AT1G48750
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.187 |
-0.010 |
| 81 |
244985_at |
ATCG00810
|
RPL22, ribosomal protein L22 |
-1.234 |
-0.041 |
| 82 |
245025_at |
ATCG00130
|
ATPF |
-1.114 |
-0.080 |
| 83 |
244981_at |
ATCG00770
|
RPS8, ribosomal protein S8 |
-1.106 |
-0.069 |
| 84 |
244980_at |
ATCG00760
|
RPL36, ribosomal protein L36 |
-1.031 |
-0.057 |
| 85 |
244966_at |
ATCG00600
|
PETG |
-1.009 |
-0.016 |
| 86 |
245008_at |
ATCG00360
|
YCF3 |
-0.910 |
-0.104 |
| 87 |
245049_at |
ATCG00050
|
RPS16, ribosomal protein S16 |
-0.904 |
-0.066 |
| 88 |
244970_at |
ATCG00660
|
RPL20, ribosomal protein L20 |
-0.898 |
-0.068 |
| 89 |
244939_at |
ATCG00065
|
RPS12, RIBOSOMAL PROTEIN S12, RPS12A, ribosomal protein S12A |
-0.850 |
-0.152 |
| 90 |
244983_at |
ATCG00790
|
RPL16, ribosomal protein L16 |
-0.832 |
-0.066 |
| 91 |
245017_at |
ATCG00510
|
PSAI, photsystem I subunit I |
-0.812 |
-0.045 |
| 92 |
245016_at |
ATCG00500
|
ACCD, acetyl-CoA carboxylase carboxyl transferase subunit beta |
-0.798 |
-0.106 |
| 93 |
244932_at |
ATCG01060
|
PSAC |
-0.796 |
0.012 |
| 94 |
245011_at |
ATCG00430
|
PSBG, photosystem II reaction center protein G |
-0.784 |
-0.081 |
| 95 |
244965_at |
ATCG00590
|
ORF31 |
-0.760 |
0.093 |
| 96 |
244936_at |
ATCG01100
|
NDHA |
-0.741 |
-0.007 |
| 97 |
244982_at |
ATCG00780
|
RPL14, ribosomal protein L14 |
-0.733 |
-0.079 |
| 98 |
244998_at |
ATCG00180
|
RPOC1 |
-0.721 |
-0.072 |
| 99 |
244984_at |
ATCG00800
|
[RESISTANCE TO PSEUDOMONAS SYRINGAE 3 (RPS3)] |
-0.715 |
0.001 |
| 100 |
244933_at |
ATCG01070
|
NDHE |
-0.709 |
-0.001 |
| 101 |
245021_at |
ATCG00550
|
PSBJ, photosystem II reaction center protein J |
-0.706 |
0.005 |
| 102 |
244975_at |
ATCG00710
|
PSBH, photosystem II reaction center protein H |
-0.704 |
-0.043 |
| 103 |
245003_at |
ATCG00280
|
PSBC, photosystem II reaction center protein C |
-0.702 |
-0.067 |
| 104 |
245010_at |
ATCG00420
|
NDHJ, NADH dehydrogenase subunit J |
-0.693 |
-0.055 |
| 105 |
245026_at |
ATCG00140
|
ATPH |
-0.657 |
-0.060 |
| 106 |
245019_at |
ATCG00530
|
YCF10 |
-0.646 |
-0.036 |
| 107 |
244963_at |
ATCG00570
|
PSBF, photosystem II reaction center protein F |
-0.639 |
-0.011 |
| 108 |
249271_at |
AT5G41790
|
CIP1, COP1-interactive protein 1 |
-0.636 |
-0.012 |
| 109 |
244934_at |
ATCG01080
|
NDHG |
-0.634 |
-0.019 |
| 110 |
244996_at |
ATCG00160
|
RPS2, ribosomal protein S2 |
-0.632 |
-0.079 |
| 111 |
244967_at |
ATCG00630
|
PSAJ |
-0.603 |
0.039 |
| 112 |
245015_at |
ATCG00490
|
RBCL |
-0.601 |
-0.082 |
| 113 |
244986_at |
ATCG00820
|
RPS19, ribosomal protein S19 |
-0.601 |
0.002 |
| 114 |
257334_at |
ATMG01370
|
ORF111D |
-0.590 |
0.043 |
| 115 |
244964_at |
ATCG00580
|
PSBE, photosystem II reaction center protein E |
-0.584 |
-0.011 |
| 116 |
244979_at |
ATCG00750
|
RPS11, ribosomal protein S11 |
-0.566 |
-0.022 |
| 117 |
244960_at |
ATCG01020
|
RPL32, ribosomal protein L32 |
-0.553 |
0.058 |
| 118 |
244997_at |
ATCG00170
|
RPOC2 |
-0.551 |
-0.132 |
| 119 |
245004_at |
ATCG00300
|
YCF9 |
-0.544 |
-0.018 |
| 120 |
245009_at |
ATCG00380
|
RPS4, chloroplast ribosomal protein S4 |
-0.538 |
-0.049 |
| 121 |
245018_at |
ATCG00520
|
YCF4 |
-0.527 |
-0.070 |
| 122 |
245005_at |
ATCG00330
|
RPS14, chloroplast ribosomal protein S14 |
-0.503 |
-0.041 |
| 123 |
245024_at |
ATCG00120
|
ATPA, ATP synthase subunit alpha |
-0.502 |
-0.021 |
| 124 |
244969_at |
ATCG00650
|
RPS18, ribosomal protein S18 |
-0.502 |
-0.002 |
| 125 |
244937_at |
ATCG01110
|
NDHH, NAD(P)H dehydrogenase subunit H |
-0.496 |
-0.082 |
| 126 |
245014_at |
ATCG00480
|
ATPB, ATP synthase subunit beta, PB, ATP synthase subunit beta |
-0.494 |
-0.028 |
| 127 |
258863_at |
AT3G03300
|
ATDCL2, DICER-LIKE 2, DCL2, dicer-like 2 |
-0.489 |
0.040 |
| 128 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.486 |
0.073 |
| 129 |
244935_at |
ATCG01090
|
NDHI |
-0.464 |
-0.017 |
| 130 |
245006_at |
ATCG00340
|
PSAB |
-0.464 |
-0.015 |
| 131 |
255703_at |
AT4G00040
|
[Chalcone and stilbene synthase family protein] |
-0.455 |
-0.042 |
| 132 |
261845_at |
AT1G15960
|
ATNRAMP6, NRAMP6, NRAMP metal ion transporter 6 |
-0.454 |
0.122 |
| 133 |
259573_at |
AT1G20390
|
[gypsy-like retrotransposon family, has a 1.5e-251 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
-0.449 |
-0.216 |
| 134 |
249542_at |
AT5G38140
|
NF-YC12, nuclear factor Y, subunit C12 |
-0.437 |
-0.078 |
| 135 |
244962_at |
ATCG01050
|
NDHD |
-0.433 |
-0.049 |
| 136 |
244976_at |
ATCG00720
|
PETB, photosynthetic electron transfer B |
-0.432 |
-0.030 |
| 137 |
251347_at |
AT3G61010
|
[Ferritin/ribonucleotide reductase-like family protein] |
-0.425 |
0.025 |
| 138 |
252208_at |
AT3G50380
|
[INVOLVED IN: protein localization] |
-0.424 |
0.002 |
| 139 |
266971_at |
AT2G39580
|
unknown |
-0.424 |
-0.040 |
| 140 |
249006_at |
AT5G44660
|
unknown |
-0.418 |
-0.053 |
| 141 |
254247_at |
AT4G23260
|
CRK18, cysteine-rich RLK (RECEPTOR-like protein kinase) 18 |
-0.414 |
0.058 |
| 142 |
245020_at |
ATCG00540
|
PETA, photosynthetic electron transfer A |
-0.411 |
-0.033 |
| 143 |
250228_at |
AT5G13840
|
CCS52B, cell cycle switch protein 52 B, FZR3, FIZZY-related 3 |
-0.405 |
0.073 |
| 144 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
-0.405 |
-0.022 |
| 145 |
244961_at |
ATCG01040
|
YCF5 |
-0.403 |
-0.026 |
| 146 |
249144_at |
AT5G43270
|
SPL2, squamosa promoter binding protein-like 2 |
-0.401 |
-0.070 |
| 147 |
265902_at |
AT2G25590
|
[Plant Tudor-like protein] |
-0.401 |
0.059 |
| 148 |
244968_at |
ATCG00640
|
RPL33, ribosomal protein L33 |
-0.387 |
-0.001 |
| 149 |
257971_at |
AT3G27530
|
GC6, golgin candidate 6, MAG4, MAIGO 4 |
-0.384 |
-0.083 |
| 150 |
248727_at |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
-0.381 |
-0.058 |
| 151 |
245050_at |
ATCG00070
|
PSBK, photosystem II reaction center protein K precursor |
-0.379 |
-0.012 |
| 152 |
244971_at |
ATCG00670
|
CLPP1, CASEINOLYTIC PROTEASE P 1, PCLPP, plastid-encoded CLP P |
-0.366 |
-0.040 |
| 153 |
244906_at |
ATMG00690
|
ORF240A |
-0.358 |
0.067 |
| 154 |
246897_at |
AT5G25560
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.352 |
-0.006 |
| 155 |
256734_at |
AT3G29390
|
RIK, RS2-interacting KH protein |
-0.352 |
-0.069 |
| 156 |
264166_at |
AT1G65370
|
[TRAF-like family protein] |
-0.349 |
0.092 |
| 157 |
244978_at |
ATCG00740
|
RPOA, RNA polymerase subunit alpha |
-0.346 |
-0.079 |
| 158 |
249808_at |
AT5G23890
|
[LOCATED IN: mitochondrion, chloroplast thylakoid membrane, chloroplast, plastid, chloroplast envelope] |
-0.336 |
-0.036 |
| 159 |
252537_at |
AT3G45710
|
[Major facilitator superfamily protein] |
-0.336 |
-0.130 |