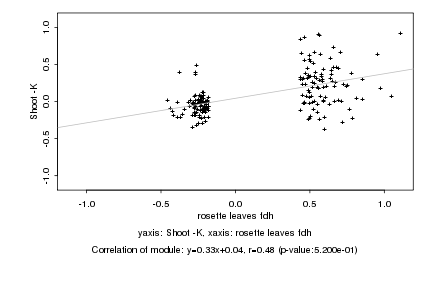
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
rosette leaves fdh |
Log2 signal ratio
Shoot -K |
| 1 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
1.106 |
0.921 |
| 2 |
250500_at |
AT5G09530
|
PELPK1, Pro-Glu-Leu|Ile|Val-Pro-Lys 1, PRP10, proline-rich protein 10 |
1.044 |
0.070 |
| 3 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.974 |
0.178 |
| 4 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
0.952 |
0.635 |
| 5 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
0.851 |
0.029 |
| 6 |
246831_at |
AT5G26340
|
ATSTP13, SUGAR TRANSPORT PROTEIN 13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
0.847 |
0.307 |
| 7 |
251065_at |
AT5G01870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.807 |
0.044 |
| 8 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.785 |
-0.223 |
| 9 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
0.779 |
0.383 |
| 10 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
0.763 |
-0.102 |
| 11 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.747 |
0.229 |
| 12 |
267008_at |
AT2G39350
|
ABCG1, ATP-binding cassette G1 |
0.741 |
0.203 |
| 13 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.720 |
0.235 |
| 14 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
0.719 |
-0.279 |
| 15 |
245076_at |
AT2G23170
|
GH3.3 |
0.706 |
0.012 |
| 16 |
258209_at |
AT3G14060
|
unknown |
0.705 |
0.672 |
| 17 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
0.688 |
0.020 |
| 18 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
0.686 |
0.450 |
| 19 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.675 |
0.463 |
| 20 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.667 |
0.270 |
| 21 |
248218_at |
AT5G53710
|
unknown |
0.662 |
0.215 |
| 22 |
263931_at |
AT2G36220
|
unknown |
0.661 |
0.004 |
| 23 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.654 |
0.466 |
| 24 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.652 |
0.734 |
| 25 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.649 |
0.273 |
| 26 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.642 |
0.419 |
| 27 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.640 |
0.366 |
| 28 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.637 |
0.316 |
| 29 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.633 |
0.590 |
| 30 |
257679_at |
AT3G20470
|
ATGRP-5, ATGRP5, GRP-5, GLYCINE-RICH PROTEIN 5, GRP5, glycine-rich protein 5 |
0.631 |
-0.032 |
| 31 |
254243_at |
AT4G23210
|
CRK13, cysteine-rich RLK (RECEPTOR-like protein kinase) 13 |
0.609 |
0.206 |
| 32 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
0.602 |
0.065 |
| 33 |
251673_at |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
0.593 |
-0.373 |
| 34 |
248163_at |
AT5G54510
|
DFL1, DWARF IN LIGHT 1, GH3.6, Gretchen Hagen3.6 |
0.592 |
-0.207 |
| 35 |
256933_at |
AT3G22600
|
LTPG5, glycosylphosphatidylinositol-anchored lipid protein transfer 5 |
0.590 |
0.438 |
| 36 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
0.590 |
0.201 |
| 37 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
0.588 |
0.012 |
| 38 |
247492_at |
AT5G61890
|
[Integrase-type DNA-binding superfamily protein] |
0.586 |
0.025 |
| 39 |
267181_at |
AT2G37760
|
AKR4C8, Aldo-keto reductase family 4 member C8 |
0.580 |
0.281 |
| 40 |
262887_at |
AT1G14780
|
[MAC/Perforin domain-containing protein] |
0.580 |
0.301 |
| 41 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.573 |
0.338 |
| 42 |
255596_at |
AT4G01720
|
AtWRKY47, WRKY47 |
0.572 |
0.375 |
| 43 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
0.568 |
0.644 |
| 44 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
0.566 |
0.077 |
| 45 |
264580_at |
AT1G05340
|
unknown |
0.564 |
0.892 |
| 46 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
0.562 |
-0.229 |
| 47 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.561 |
0.294 |
| 48 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.556 |
0.179 |
| 49 |
245840_at |
AT1G58420
|
[Uncharacterised conserved protein UCP031279] |
0.553 |
0.906 |
| 50 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
0.552 |
0.179 |
| 51 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.551 |
0.200 |
| 52 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.549 |
-0.143 |
| 53 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.549 |
-0.040 |
| 54 |
250670_at |
AT5G06860
|
ATPGIP1, POLYGALACTURONASE INHIBITING PROTEIN 1, PGIP1, polygalacturonase inhibiting protein 1 |
0.540 |
0.321 |
| 55 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
0.532 |
0.399 |
| 56 |
260744_at |
AT1G15010
|
unknown |
0.531 |
0.666 |
| 57 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
0.531 |
0.011 |
| 58 |
266246_at |
AT2G27690
|
CYP94C1, cytochrome P450, family 94, subfamily C, polypeptide 1 |
0.529 |
0.255 |
| 59 |
256937_at |
AT3G22620
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.527 |
0.348 |
| 60 |
247177_at |
AT5G65300
|
unknown |
0.522 |
0.519 |
| 61 |
252604_at |
AT3G45060
|
ATNRT2.6, ARABIDOPSIS THALIANA HIGH AFFINITY NITRATE TRANSPORTER 2.6, NRT2.6, high affinity nitrate transporter 2.6 |
0.519 |
-0.095 |
| 62 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
0.517 |
0.000 |
| 63 |
253173_at |
AT4G35110
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.515 |
0.196 |
| 64 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.515 |
0.267 |
| 65 |
249719_at |
AT5G35735
|
[Auxin-responsive family protein] |
0.514 |
0.263 |
| 66 |
248368_at |
AT5G51950
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
0.505 |
0.071 |
| 67 |
247013_at |
AT5G67480
|
ATBT4, BT4, BTB and TAZ domain protein 4 |
0.503 |
-0.195 |
| 68 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.499 |
0.345 |
| 69 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.499 |
0.545 |
| 70 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.496 |
0.130 |
| 71 |
247951_at |
AT5G57240
|
ORP4C, OSBP(oxysterol binding protein)-related protein 4C |
0.496 |
0.064 |
| 72 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.494 |
0.627 |
| 73 |
253309_at |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
0.492 |
-0.218 |
| 74 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
0.491 |
-0.024 |
| 75 |
257644_at |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
0.491 |
0.577 |
| 76 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
0.490 |
0.333 |
| 77 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
0.489 |
0.356 |
| 78 |
251633_at |
AT3G57460
|
[catalytics] |
0.486 |
0.152 |
| 79 |
262947_at |
AT1G75750
|
GASA1, GAST1 protein homolog 1 |
0.485 |
-0.230 |
| 80 |
266017_at |
AT2G18690
|
unknown |
0.480 |
0.311 |
| 81 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
0.478 |
0.450 |
| 82 |
258037_at |
AT3G21230
|
4CL5, 4-coumarate:CoA ligase 5 |
0.477 |
0.077 |
| 83 |
246018_at |
AT5G10695
|
unknown |
0.466 |
0.382 |
| 84 |
259385_at |
AT1G13470
|
unknown |
0.464 |
0.242 |
| 85 |
248509_at |
AT5G50335
|
unknown |
0.462 |
-0.014 |
| 86 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
0.462 |
0.557 |
| 87 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
0.461 |
-0.005 |
| 88 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
0.459 |
0.873 |
| 89 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
0.452 |
0.322 |
| 90 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
0.451 |
-0.023 |
| 91 |
245252_at |
AT4G17500
|
ATERF-1, ethylene responsive element binding factor 1, ERF-1, ethylene responsive element binding factor 1 |
0.449 |
0.299 |
| 92 |
245731_at |
AT1G73500
|
ATMKK9, MKK9, MAP kinase kinase 9 |
0.448 |
0.092 |
| 93 |
263536_at |
AT2G25000
|
ATWRKY60, WRKY60, WRKY DNA-binding protein 60 |
0.444 |
0.242 |
| 94 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.441 |
0.657 |
| 95 |
265853_at |
AT2G42360
|
[RING/U-box superfamily protein] |
0.434 |
0.838 |
| 96 |
248551_at |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
0.434 |
0.298 |
| 97 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.433 |
0.328 |
| 98 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
0.431 |
-0.112 |
| 99 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
-0.461 |
0.016 |
| 100 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
-0.441 |
-0.087 |
| 101 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
-0.423 |
-0.126 |
| 102 |
267377_at |
AT2G26250
|
FDH, FIDDLEHEAD, KCS10, 3-ketoacyl-CoA synthase 10 |
-0.420 |
-0.176 |
| 103 |
260297_at |
AT1G80280
|
[alpha/beta-Hydrolases superfamily protein] |
-0.392 |
-0.204 |
| 104 |
251750_at |
AT3G55710
|
[UDP-Glycosyltransferase superfamily protein] |
-0.390 |
-0.003 |
| 105 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
-0.382 |
0.399 |
| 106 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.373 |
-0.208 |
| 107 |
254125_at |
AT4G24670
|
TAR2, tryptophan aminotransferase related 2 |
-0.359 |
-0.168 |
| 108 |
261660_at |
AT1G18370
|
ATNACK1, ARABIDOPSIS NPK1-ACTIVATING KINESIN 1, HIK, HINKEL, NACK1, NPK1-ACTIVATING KINESIN 1 |
-0.343 |
-0.106 |
| 109 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
-0.318 |
-0.002 |
| 110 |
248807_at |
AT5G47500
|
PME5, pectin methylesterase 5 |
-0.309 |
-0.059 |
| 111 |
259042_at |
AT3G03450
|
RGL2, RGA-like 2 |
-0.307 |
0.025 |
| 112 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
-0.294 |
-0.026 |
| 113 |
261330_at |
AT1G44900
|
ATMCM2, MCM2, MINICHROMOSOME MAINTENANCE 2 |
-0.294 |
-0.350 |
| 114 |
260141_at |
AT1G66350
|
RGL, RGL1, RGA-like 1 |
-0.293 |
-0.181 |
| 115 |
251605_at |
AT3G57830
|
[Leucine-rich repeat protein kinase family protein] |
-0.286 |
-0.038 |
| 116 |
266655_at |
AT2G25880
|
AtAUR2, ataurora2, AUR2, ataurora2 |
-0.284 |
-0.004 |
| 117 |
251586_at |
AT3G58070
|
GIS, GLABROUS INFLORESCENCE STEMS |
-0.280 |
0.076 |
| 118 |
245576_at |
AT4G14770
|
ATTCX2, TESMIN/TSO1-LIKE CXC 2, TCX2, TESMIN/TSO1-like CXC 2 |
-0.279 |
-0.070 |
| 119 |
262854_at |
AT1G20870
|
[HSP20-like chaperones superfamily protein] |
-0.279 |
-0.029 |
| 120 |
255675_at |
AT4G00480
|
ATMYC1, myc1 |
-0.278 |
-0.136 |
| 121 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.278 |
-0.181 |
| 122 |
262907_at |
AT1G59720
|
CRR28, CHLORORESPIRATORY REDUCTION28 |
-0.277 |
-0.052 |
| 123 |
262238_at |
AT1G48300
|
DGAT3, diacylglycerol acyltransferase 3 |
-0.276 |
-0.081 |
| 124 |
256864_at |
AT3G23890
|
ATTOPII, TOPII, topoisomerase II |
-0.274 |
-0.056 |
| 125 |
251826_at |
AT3G55110
|
ABCG18, ATP-binding cassette G18 |
-0.274 |
0.087 |
| 126 |
256125_at |
AT1G18250
|
ATLP-1 |
-0.274 |
-0.158 |
| 127 |
247599_at |
AT5G60880
|
BASL, BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE |
-0.273 |
0.021 |
| 128 |
251886_at |
AT3G54260
|
TBL36, TRICHOME BIREFRINGENCE-LIKE 36 |
-0.271 |
-0.034 |
| 129 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
-0.270 |
-0.179 |
| 130 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
-0.270 |
0.397 |
| 131 |
265913_at |
AT2G25625
|
unknown |
-0.269 |
0.377 |
| 132 |
248696_at |
AT5G48360
|
[Actin-binding FH2 (formin homology 2) family protein] |
-0.268 |
-0.139 |
| 133 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
-0.267 |
-0.313 |
| 134 |
259133_at |
AT3G05400
|
[Major facilitator superfamily protein] |
-0.262 |
0.489 |
| 135 |
257115_at |
AT3G20150
|
[Kinesin motor family protein] |
-0.260 |
-0.024 |
| 136 |
255265_at |
AT4G05190
|
ATK5, kinesin 5 |
-0.255 |
-0.103 |
| 137 |
248020_at |
AT5G56490
|
AtGulLO4, GulLO4, L -gulono-1,4-lactone ( L -GulL) oxidase 4 |
-0.254 |
0.028 |
| 138 |
252089_at |
AT3G52110
|
unknown |
-0.254 |
0.017 |
| 139 |
248891_at |
AT5G46280
|
MCM3, MINICHROMOSOME MAINTENANCE 3 |
-0.254 |
-0.286 |
| 140 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
-0.248 |
-0.186 |
| 141 |
256077_at |
AT1G18090
|
[5'-3' exonuclease family protein] |
-0.245 |
-0.205 |
| 142 |
263441_at |
AT2G28620
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.245 |
-0.003 |
| 143 |
267205_at |
AT2G30820
|
unknown |
-0.245 |
0.082 |
| 144 |
245739_at |
AT1G44110
|
CYCA1;1, Cyclin A1;1 |
-0.241 |
0.020 |
| 145 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
-0.240 |
-0.058 |
| 146 |
254096_at |
AT4G25150
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.239 |
0.075 |
| 147 |
265171_at |
AT1G23790
|
unknown |
-0.237 |
-0.024 |
| 148 |
246315_at |
AT3G56870
|
unknown |
-0.237 |
-0.064 |
| 149 |
250413_at |
AT5G11160
|
APT5, adenine phosphoribosyltransferase 5 |
-0.236 |
-0.117 |
| 150 |
263480_at |
AT2G04032
|
ZIP7, zinc transporter 7 precursor |
-0.236 |
0.032 |
| 151 |
247028_at |
AT5G67100
|
ICU2, INCURVATA2 |
-0.235 |
-0.115 |
| 152 |
267173_at |
AT2G37560
|
ATORC2, ORIGIN RECOGNITION COMPLEX SECOND LARGEST SUBUNIT 2, ORC2, origin recognition complex second largest subunit 2 |
-0.234 |
-0.042 |
| 153 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
-0.233 |
-0.026 |
| 154 |
246535_at |
AT5G15900
|
TBL19, TRICHOME BIREFRINGENCE-LIKE 19 |
-0.233 |
0.080 |
| 155 |
251704_at |
AT3G56360
|
unknown |
-0.233 |
0.061 |
| 156 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.232 |
0.014 |
| 157 |
255851_at |
AT1G67040
|
TRM22, TON1 Recruiting Motif 22 |
-0.232 |
-0.220 |
| 158 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.225 |
0.126 |
| 159 |
246069_at |
AT5G20220
|
[zinc knuckle (CCHC-type) family protein] |
-0.224 |
-0.140 |
| 160 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.222 |
-0.106 |
| 161 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
-0.222 |
-0.295 |
| 162 |
264550_at |
AT1G09450
|
AtHaspin, Haspin, Haspin-related gene |
-0.222 |
0.003 |
| 163 |
253403_at |
AT4G32830
|
AtAUR1, ataurora1, AUR1, ataurora1 |
-0.220 |
-0.108 |
| 164 |
248614_at |
AT5G49560
|
[Putative methyltransferase family protein] |
-0.219 |
0.130 |
| 165 |
249546_at |
AT5G38150
|
PMI15, plastid movement impaired 15 |
-0.218 |
-0.085 |
| 166 |
250806_at |
AT5G05070
|
[DHHC-type zinc finger family protein] |
-0.216 |
0.045 |
| 167 |
253514_at |
AT4G31805
|
POLAR, POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION |
-0.216 |
-0.012 |
| 168 |
251271_at |
AT3G62050
|
[Putative endonuclease or glycosyl hydrolase] |
-0.215 |
0.087 |
| 169 |
257801_at |
AT3G18750
|
ATWNK6, ARABIDOPSIS THALIANA WITH NO K 6, WNK6, with no lysine (K) kinase 6, ZIK5 |
-0.214 |
-0.123 |
| 170 |
258409_at |
AT3G17640
|
[Leucine-rich repeat (LRR) family protein] |
-0.214 |
-0.116 |
| 171 |
256832_at |
AT3G22880
|
ARLIM15, ARABIDOPSIS HOMOLOG OF LILY MESSAGES INDUCED AT MEIOSIS 15, ATDMC1, ARABIDOPSIS THALIANA DISRUPTION OF MEIOTIC CONTROL 1, DMC1, DISRUPTION OF MEIOTIC CONTROL 1 |
-0.213 |
-0.076 |
| 172 |
259823_at |
AT1G66250
|
[O-Glycosyl hydrolases family 17 protein] |
-0.213 |
-0.105 |
| 173 |
262737_at |
AT1G28560
|
SRD2, SHOOT REDIFFERENTIATION DEFECTIVE 2 |
-0.212 |
-0.021 |
| 174 |
266244_at |
AT2G27740
|
unknown |
-0.210 |
-0.056 |
| 175 |
253270_at |
AT4G34160
|
CYCD3, CYCD3;1, CYCLIN D3;1 |
-0.208 |
-0.212 |
| 176 |
257120_at |
AT3G20200
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.207 |
0.004 |
| 177 |
251714_at |
AT3G56370
|
[Leucine-rich repeat protein kinase family protein] |
-0.205 |
-0.264 |
| 178 |
250560_at |
AT5G08020
|
ATRPA70B, ARABIDOPSIS THALIANA RPA70-KDA SUBUNIT B, RPA70B, RPA70-kDa subunit B |
-0.204 |
-0.084 |
| 179 |
252025_at |
AT3G52900
|
unknown |
-0.204 |
-0.147 |
| 180 |
262619_at |
AT1G06550
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
-0.203 |
-0.068 |
| 181 |
259945_at |
AT1G71460
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
-0.201 |
-0.035 |
| 182 |
266007_at |
AT2G37380
|
MAKR3, MEMBRANE-ASSOCIATED KINASE REGULATOR 3 |
-0.200 |
-0.085 |
| 183 |
254443_at |
AT4G21070
|
ATBRCA1, ARABIDOPSIS THALIANA BREAST CANCER SUSCEPTIBILITY1, BRCA1, breast cancer susceptibility1 |
-0.199 |
-0.029 |
| 184 |
252361_at |
AT3G48490
|
unknown |
-0.198 |
-0.057 |
| 185 |
259851_at |
AT1G72250
|
[Di-glucose binding protein with Kinesin motor domain] |
-0.198 |
-0.028 |
| 186 |
262438_at |
AT1G47410
|
unknown |
-0.197 |
0.003 |
| 187 |
256320_at |
AT3G12170
|
[Chaperone DnaJ-domain superfamily protein] |
-0.194 |
-0.083 |
| 188 |
246900_at |
AT5G25620
|
AtYUC6, YUC6, YUCCA6 |
-0.190 |
0.007 |
| 189 |
247118_at |
AT5G65890
|
ACR1, ACT domain repeat 1 |
-0.188 |
0.019 |
| 190 |
247034_at |
AT5G67260
|
CYCD3;2, CYCLIN D3;2 |
-0.187 |
-0.065 |
| 191 |
264319_at |
AT1G04110
|
AtSDD1, SDD1, STOMATAL DENSITY AND DISTRIBUTION 1 |
-0.186 |
-0.061 |
| 192 |
265383_at |
AT2G16780
|
MSI02, MSI2, MULTICOPY SUPPRESSOR OF IRA1 2, NFC02, NFC2, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP C 2 |
-0.186 |
-0.121 |
| 193 |
260364_at |
AT1G70560
|
CKRC1, cytokinin induced root curling 1, SAV3, SHADE AVOIDANCE 3, TAA1, tryptophan aminotransferase of Arabidopsis 1, WEI8, WEAK ETHYLENE INSENSITIVE 8 |
-0.184 |
-0.007 |
| 194 |
251733_at |
AT3G56240
|
CCH, copper chaperone |
-0.184 |
0.064 |
| 195 |
246783_at |
AT5G27360
|
SFP2 |
-0.184 |
-0.208 |