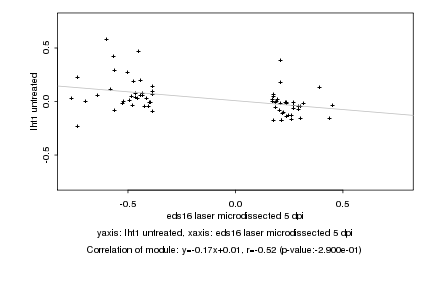
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
eds16 laser microdissected 5 dpi |
Log2 signal ratio
lht1 untreated |
| 1 |
266025_at |
AT2G05960
|
[copia-like retrotransposon family, has a 6.2e-200 P-value blast match to gb|AAG52949.1| gag/pol polyprotein (Endovir1-1) (Arabidopsis thaliana) (Ty1_Copia-family)] |
0.447 |
-0.030 |
| 2 |
267416_at |
AT2G34980
|
SETH1 |
0.435 |
-0.156 |
| 3 |
246143_at |
AT5G19980
|
GONST4, golgi nucleotide sugar transporter 4 |
0.389 |
0.133 |
| 4 |
248028_at |
AT5G55620
|
unknown |
0.312 |
-0.015 |
| 5 |
255905_at |
AT1G17810
|
BETA-TIP, beta-tonoplast intrinsic protein |
0.300 |
-0.155 |
| 6 |
250239_at |
AT5G13580
|
ABCG6, ATP-binding cassette G6 |
0.298 |
-0.046 |
| 7 |
267594_at |
AT2G33000
|
[ubiquitin-associated (UBA)/TS-N domain-containing protein-related] |
0.290 |
-0.072 |
| 8 |
265703_at |
AT2G03430
|
[Ankyrin repeat family protein] |
0.290 |
-0.038 |
| 9 |
245490_at |
AT4G16310
|
LDL3, LSD1-like 3 |
0.268 |
-0.006 |
| 10 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
0.267 |
-0.028 |
| 11 |
247524_at |
AT5G61440
|
ACHT5, atypical CYS HIS rich thioredoxin 5 |
0.266 |
-0.059 |
| 12 |
248168_at |
AT5G54570
|
BGLU41, beta glucosidase 41 |
0.258 |
-0.124 |
| 13 |
249109_at |
AT5G43700
|
ATAUX2-11, AUXIN INDUCIBLE 2-11, IAA4, indole-3-acetic acid inducible 4 |
0.257 |
-0.159 |
| 14 |
259021_at |
AT3G07540
|
[Actin-binding FH2 (formin homology 2) family protein] |
0.243 |
-0.125 |
| 15 |
252513_at |
AT3G46220
|
unknown |
0.237 |
-0.003 |
| 16 |
246663_at |
AT5G35300
|
unknown |
0.234 |
-0.136 |
| 17 |
245322_at |
AT4G14815
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.233 |
-0.014 |
| 18 |
255244_at |
AT4G05620
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.229 |
-0.006 |
| 19 |
264127_at |
AT1G79250
|
AGC1.7, AGC kinase 1.7 |
0.220 |
-0.095 |
| 20 |
256668_at |
AT3G32190
|
unknown |
0.218 |
-0.105 |
| 21 |
254410_at |
AT4G21410
|
CRK29, cysteine-rich RLK (RECEPTOR-like protein kinase) 29 |
0.213 |
-0.171 |
| 22 |
246919_at |
AT5G25460
|
DGR2, DUF642 L-GalL responsive gene 2 |
0.211 |
-0.174 |
| 23 |
246477_at |
AT5G16770
|
AtMYB9, myb domain protein 9, MYB9, myb domain protein 9 |
0.209 |
-0.012 |
| 24 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
0.208 |
0.391 |
| 25 |
258545_at |
AT3G07050
|
NSN1, nucleostemin-like 1 |
0.206 |
0.187 |
| 26 |
257309_at |
AT3G28150
|
AXY4L, ALTERED XYLOGLUCAN 4-LIKE, TBL22, TRICHOME BIREFRINGENCE-LIKE 22 |
0.203 |
-0.082 |
| 27 |
255864_at |
AT2G30320
|
[Pseudouridine synthase family protein] |
0.195 |
0.019 |
| 28 |
256479_at |
AT1G33400
|
TPR9, tetratricopeptide repeat 9 |
0.188 |
0.006 |
| 29 |
266773_at |
AT2G29040
|
[Exostosin family protein] |
0.184 |
-0.003 |
| 30 |
256456_at |
AT1G75180
|
[Erythronate-4-phosphate dehydrogenase family protein] |
0.183 |
-0.054 |
| 31 |
267620_at |
AT2G39640
|
[glycosyl hydrolase family 17 protein] |
0.177 |
-0.174 |
| 32 |
254060_at |
AT4G25350
|
SHB1, SHORT HYPOCOTYL UNDER BLUE1 |
0.177 |
0.048 |
| 33 |
249386_at |
AT5G40060
|
[Disease resistance protein (NBS-LRR class) family] |
0.174 |
0.066 |
| 34 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
0.172 |
0.002 |
| 35 |
250223_at |
AT5G14070
|
ROXY2 |
0.171 |
0.025 |
| 36 |
254429_at |
AT4G21105
|
[cytochrome-c oxidases] |
-0.766 |
0.035 |
| 37 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.738 |
0.228 |
| 38 |
254431_at |
AT4G20840
|
[FAD-binding Berberine family protein] |
-0.737 |
-0.229 |
| 39 |
252618_at |
AT3G45140
|
ATLOX2, ARABIODOPSIS THALIANA LIPOXYGENASE 2, LOX2, lipoxygenase 2 |
-0.701 |
0.008 |
| 40 |
259706_at |
AT1G77540
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.642 |
0.057 |
| 41 |
267345_at |
AT2G44240
|
unknown |
-0.603 |
0.586 |
| 42 |
266696_at |
AT2G19680
|
[Mitochondrial ATP synthase subunit G protein] |
-0.583 |
0.119 |
| 43 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
-0.569 |
0.421 |
| 44 |
255302_at |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
-0.567 |
-0.076 |
| 45 |
247314_at |
AT5G64000
|
ATSAL2, SAL2 |
-0.564 |
0.297 |
| 46 |
265226_at |
AT2G28430
|
unknown |
-0.527 |
-0.011 |
| 47 |
245626_at |
AT1G56700
|
[Peptidase C15, pyroglutamyl peptidase I-like] |
-0.524 |
0.003 |
| 48 |
267300_at |
AT2G30140
|
UGT87A2, UDP-glucosyl transferase 87A2 |
-0.503 |
0.277 |
| 49 |
245279_at |
AT4G17270
|
[Mo25 family protein] |
-0.497 |
0.017 |
| 50 |
248951_at |
AT5G45550
|
MOB1-like, MOB1-like |
-0.485 |
0.048 |
| 51 |
249834_at |
AT5G23440
|
FTRA1, ferredoxin/thioredoxin reductase subunit A (variable subunit) 1 |
-0.482 |
-0.034 |
| 52 |
248321_at |
AT5G52740
|
[Copper transport protein family] |
-0.478 |
0.189 |
| 53 |
249523_at |
AT5G38630
|
ACYB-1, cytochrome B561-1, CYB-1, cytochrome B561-1 |
-0.467 |
0.075 |
| 54 |
262426_s_at |
AT1G47640
|
unknown |
-0.465 |
0.046 |
| 55 |
245175_at |
AT2G47470
|
ATPDI11, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 11, ATPDIL2-1, PDI-LIKE 2-1, MEE30, MATERNAL EFFECT EMBRYO ARREST 30, PDI11, PROTEIN DISULFIDE ISOMERASE 11, UNE5, UNFERTILIZED EMBRYO SAC 5 |
-0.459 |
0.036 |
| 56 |
264434_at |
AT1G10340
|
[Ankyrin repeat family protein] |
-0.451 |
0.468 |
| 57 |
266093_at |
AT2G37990
|
[ribosome biogenesis regulatory protein (RRS1) family protein] |
-0.443 |
0.060 |
| 58 |
255243_at |
AT4G05590
|
unknown |
-0.442 |
0.204 |
| 59 |
262876_at |
AT1G64750
|
ATDSS1(I), deletion of SUV3 suppressor 1(I), DSS1(I), deletion of SUV3 suppressor 1(I) |
-0.435 |
0.081 |
| 60 |
264406_at |
AT1G10290
|
ADL6, dynamin-like protein 6, DRP2A, DYNAMIN-RELATED PROTEIN 2A |
-0.434 |
0.062 |
| 61 |
251561_at |
AT3G57870
|
AHUS5, ATSCE1, SUMO CONJUGATION ENZYME 1, EMB1637, EMBRYO DEFECTIVE 1637, SCE1, sumo conjugation enzyme 1, SCE1A, SUMO CONJUGATING ENZYME 1A |
-0.426 |
-0.039 |
| 62 |
250042_at |
AT5G18420
|
unknown |
-0.418 |
0.033 |
| 63 |
253990_at |
AT4G26160
|
ACHT1, atypical CYS HIS rich thioredoxin 1 |
-0.406 |
-0.043 |
| 64 |
253336_at |
AT4G33250
|
ATTIF3K1, EIF3K, eukaryotic translation initiation factor 3K, TIF3K1 |
-0.400 |
-0.004 |
| 65 |
265855_at |
AT2G42390
|
[protein kinase C substrate, heavy chain-related] |
-0.397 |
-0.006 |
| 66 |
257893_at |
AT3G17000
|
UBC32, ubiquitin-conjugating enzyme 32 |
-0.388 |
0.073 |
| 67 |
246305_at |
AT3G51890
|
CLC3, clathrin light chain 3 |
-0.387 |
0.146 |
| 68 |
263426_at |
AT2G31570
|
ATGPX2, glutathione peroxidase 2, GPX2, glutathione peroxidase 2 |
-0.386 |
0.097 |
| 69 |
254356_at |
AT4G22190
|
unknown |
-0.386 |
-0.092 |