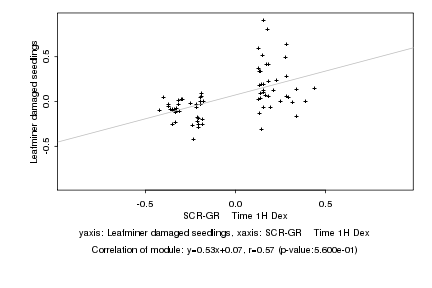
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
SCR-GR Time 1H Dex |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.437 |
0.156 |
| 2 |
253963_at |
AT4G26470
|
[Calcium-binding EF-hand family protein] |
0.386 |
0.004 |
| 3 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
0.336 |
-0.167 |
| 4 |
245566_at |
AT4G14610
|
[pseudogene] |
0.334 |
0.136 |
| 5 |
247061_at |
AT5G66780
|
unknown |
0.314 |
-0.007 |
| 6 |
249364_at |
AT5G40590
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.292 |
0.056 |
| 7 |
263565_at |
AT2G15390
|
atfut4, FUT4, fucosyltransferase 4 |
0.282 |
0.638 |
| 8 |
246600_at |
AT5G14930
|
SAG101, senescence-associated gene 101 |
0.281 |
0.288 |
| 9 |
253322_at |
AT4G33980
|
unknown |
0.279 |
0.060 |
| 10 |
245363_at |
AT4G15120
|
[VQ motif-containing protein] |
0.273 |
0.499 |
| 11 |
259008_at |
AT3G09390
|
ATMT-1, ARABIDOPSIS THALIANA METALLOTHIONEIN-1, ATMT-K, ARABIDOPSIS THALIANA METALLOTHIONEIN-K, MT2A, metallothionein 2A |
0.246 |
0.004 |
| 12 |
247913_at |
AT5G57510
|
unknown |
0.225 |
0.239 |
| 13 |
254559_at |
AT4G19200
|
[proline-rich family protein] |
0.211 |
0.134 |
| 14 |
248570_at |
AT5G49780
|
[Leucine-rich repeat protein kinase family protein] |
0.193 |
-0.057 |
| 15 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
0.182 |
0.417 |
| 16 |
261125_at |
AT1G04990
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
0.181 |
0.226 |
| 17 |
248931_at |
AT5G46040
|
[Major facilitator superfamily protein] |
0.179 |
0.060 |
| 18 |
266246_at |
AT2G27690
|
CYP94C1, cytochrome P450, family 94, subfamily C, polypeptide 1 |
0.177 |
0.814 |
| 19 |
251200_at |
AT3G63010
|
ATGID1B, GID1B, GA INSENSITIVE DWARF1B |
0.171 |
0.416 |
| 20 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
0.167 |
0.073 |
| 21 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
0.155 |
0.915 |
| 22 |
259874_at |
AT1G76660
|
unknown |
0.153 |
0.110 |
| 23 |
261771_at |
AT1G76150
|
ATECH2, ECH2, enoyl-CoA hydratase 2 |
0.153 |
0.192 |
| 24 |
259287_at |
AT3G11490
|
[rac GTPase activating protein] |
0.151 |
-0.059 |
| 25 |
253780_at |
AT4G28400
|
[Protein phosphatase 2C family protein] |
0.151 |
0.127 |
| 26 |
262887_at |
AT1G14780
|
[MAC/Perforin domain-containing protein] |
0.147 |
0.517 |
| 27 |
267347_at |
AT2G39950
|
unknown |
0.143 |
-0.309 |
| 28 |
246305_at |
AT3G51890
|
CLC3, clathrin light chain 3 |
0.140 |
0.192 |
| 29 |
260648_at |
AT1G08050
|
[Zinc finger (C3HC4-type RING finger) family protein] |
0.138 |
0.339 |
| 30 |
253956_at |
AT4G26700
|
ATFIM1, ARABIDOPSIS THALIANA FIMBRIN 1, FIM1, fimbrin 1 |
0.135 |
0.096 |
| 31 |
262552_at |
AT1G31350
|
KUF1, KAR-UP F-box 1 |
0.135 |
0.038 |
| 32 |
267623_at |
AT2G39650
|
unknown |
0.132 |
0.339 |
| 33 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.132 |
0.186 |
| 34 |
260284_at |
AT1G80380
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.131 |
-0.129 |
| 35 |
248111_at |
AT5G55330
|
[MBOAT (membrane bound O-acyl transferase) family protein] |
0.125 |
0.023 |
| 36 |
256763_at |
AT3G16860
|
COBL8, COBRA-like protein 8 precursor |
0.124 |
0.598 |
| 37 |
263411_at |
AT2G28710
|
[C2H2-type zinc finger family protein] |
0.124 |
0.373 |
| 38 |
255460_at |
AT4G02800
|
unknown |
-0.427 |
-0.098 |
| 39 |
263850_at |
AT2G04480
|
unknown |
-0.402 |
0.047 |
| 40 |
246557_at |
AT5G15510
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.377 |
-0.024 |
| 41 |
261384_at |
AT1G05440
|
[C-8 sterol isomerases] |
-0.374 |
-0.052 |
| 42 |
252691_at |
AT3G44050
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.363 |
-0.085 |
| 43 |
263441_at |
AT2G28620
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.354 |
-0.084 |
| 44 |
253978_at |
AT4G26660
|
unknown |
-0.354 |
-0.091 |
| 45 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
-0.351 |
-0.246 |
| 46 |
267006_at |
AT2G34190
|
[Xanthine/uracil permease family protein] |
-0.342 |
-0.082 |
| 47 |
261086_at |
AT1G17460
|
TRFL3, TRF-like 3 |
-0.337 |
-0.120 |
| 48 |
246269_at |
AT4G37110
|
[Zinc-finger domain of monoamine-oxidase A repressor R1] |
-0.336 |
-0.223 |
| 49 |
261780_at |
AT1G76310
|
CYCB2;4, CYCLIN B2;4 |
-0.330 |
-0.075 |
| 50 |
259290_at |
AT3G11520
|
CYC2, CYCLIN 2, CYCB1;3, CYCLIN B1;3 |
-0.329 |
-0.103 |
| 51 |
254233_at |
AT4G23800
|
3xHMG-box2, 3xHigh Mobility Group-box2 |
-0.320 |
-0.028 |
| 52 |
254986_at |
AT4G10640
|
IQD16, IQ-domain 16 |
-0.319 |
0.018 |
| 53 |
266008_at |
AT2G37390
|
NAKR2, SODIUM POTASSIUM ROOT DEFECTIVE 2 |
-0.316 |
-0.105 |
| 54 |
263610_at |
AT2G16230
|
[O-Glycosyl hydrolases family 17 protein] |
-0.305 |
0.031 |
| 55 |
247399_at |
AT5G62960
|
unknown |
-0.298 |
0.024 |
| 56 |
254109_at |
AT4G25240
|
SKS1, SKU5 similar 1 |
-0.252 |
-0.021 |
| 57 |
248807_at |
AT5G47500
|
PME5, pectin methylesterase 5 |
-0.240 |
-0.267 |
| 58 |
248696_at |
AT5G48360
|
[Actin-binding FH2 (formin homology 2) family protein] |
-0.238 |
-0.420 |
| 59 |
254728_at |
AT4G13690
|
unknown |
-0.220 |
-0.064 |
| 60 |
253027_at |
AT4G38150
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.217 |
-0.033 |
| 61 |
258462_at |
AT3G17350
|
unknown |
-0.215 |
-0.172 |
| 62 |
252888_at |
AT4G39210
|
APL3 |
-0.213 |
-0.222 |
| 63 |
256125_at |
AT1G18250
|
ATLP-1 |
-0.209 |
-0.255 |
| 64 |
249184_at |
AT5G43020
|
[Leucine-rich repeat protein kinase family protein] |
-0.209 |
-0.288 |
| 65 |
256675_at |
AT3G52170
|
[DNA binding] |
-0.206 |
-0.188 |
| 66 |
266672_at |
AT2G29650
|
ANTR1, anion transporter 1, PHT4;1, phosphate transporter 4;1 |
-0.198 |
-0.025 |
| 67 |
255169_x_at |
AT4G07940
|
unknown |
-0.197 |
0.007 |
| 68 |
253804_at |
AT4G28230
|
unknown |
-0.196 |
0.049 |
| 69 |
250434_at |
AT5G10390
|
H3.1, histone 3.1 |
-0.193 |
0.061 |
| 70 |
264895_at |
AT1G23100
|
[GroES-like family protein] |
-0.190 |
0.090 |
| 71 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
-0.184 |
-0.199 |
| 72 |
262738_at |
AT1G28530
|
unknown |
-0.184 |
-0.253 |
| 73 |
245236_at |
AT4G25540
|
ATMSH3, MSH3, homolog of DNA mismatch repair protein MSH3 |
-0.182 |
0.006 |