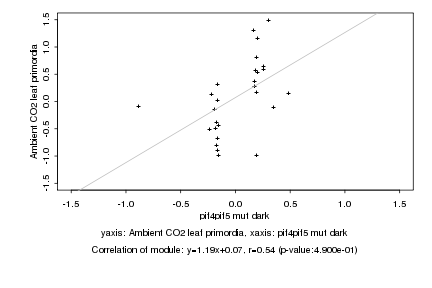
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
pif4pif5 mut dark |
Log2 signal ratio
Ambient CO2 leaf primordia |
| 1 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.477 |
0.160 |
| 2 |
260973_at |
AT1G53490
|
HEI10, homolog of human HEI10 ( Enhancer of cell Invasion No.10) |
0.347 |
-0.092 |
| 3 |
256497_at |
AT1G31580
|
CXC750, ECS1 |
0.294 |
1.504 |
| 4 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
0.256 |
0.589 |
| 5 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
0.254 |
0.643 |
| 6 |
246783_at |
AT5G27360
|
SFP2 |
0.195 |
0.540 |
| 7 |
264037_at |
AT2G03750
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.193 |
1.156 |
| 8 |
259791_at |
AT1G29700
|
[Metallo-hydrolase/oxidoreductase superfamily protein] |
0.191 |
0.825 |
| 9 |
258490_at |
AT3G02670
|
[Glycine-rich protein family] |
0.189 |
-0.977 |
| 10 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
0.188 |
0.180 |
| 11 |
248807_at |
AT5G47500
|
PME5, pectin methylesterase 5 |
0.182 |
0.573 |
| 12 |
260041_at |
AT1G68780
|
[RNI-like superfamily protein] |
0.174 |
0.285 |
| 13 |
263287_at |
AT2G36145
|
unknown |
0.173 |
0.368 |
| 14 |
251827_at |
AT3G55120
|
A11, AtCHI, CFI, CHALCONE FLAVANONE ISOMERASE, CHI, chalcone isomerase, TT5, TRANSPARENT TESTA 5 |
0.156 |
1.312 |
| 15 |
251497_at |
AT3G59060
|
PIF5, PHYTOCHROME-INTERACTING FACTOR 5, PIL6, phytochrome interacting factor 3-like 6 |
-0.893 |
-0.088 |
| 16 |
249675_at |
AT5G35940
|
[Mannose-binding lectin superfamily protein] |
-0.237 |
-0.496 |
| 17 |
266851_at |
AT2G26820
|
ATPP2-A3, phloem protein 2-A3, PP2-A3, phloem protein 2-A3 |
-0.219 |
0.144 |
| 18 |
256702_at |
AT3G30380
|
[alpha/beta-Hydrolases superfamily protein] |
-0.198 |
-0.145 |
| 19 |
265477_at |
AT2G46480
|
GAUT2, galacturonosyltransferase 2, LGT2, GALACTURONOSYLTRANSFERASE 2 |
-0.188 |
-0.482 |
| 20 |
262666_at |
AT1G14080
|
ATFUT6, FUT6, fucosyltransferase 6 |
-0.182 |
-0.793 |
| 21 |
251291_at |
AT3G61900
|
SAUR33, SMALL AUXIN UPREGULATED RNA 33 |
-0.176 |
-0.380 |
| 22 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
-0.173 |
-0.674 |
| 23 |
263580_at |
AT2G17140
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.170 |
0.019 |
| 24 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.167 |
0.319 |
| 25 |
264571_at |
AT1G05330
|
unknown |
-0.165 |
-0.886 |
| 26 |
248674_at |
AT5G48800
|
[Phototropic-responsive NPH3 family protein] |
-0.159 |
-0.433 |
| 27 |
253787_at |
AT4G28670
|
[FUNCTIONS IN: kinase activity] |
-0.156 |
-0.979 |