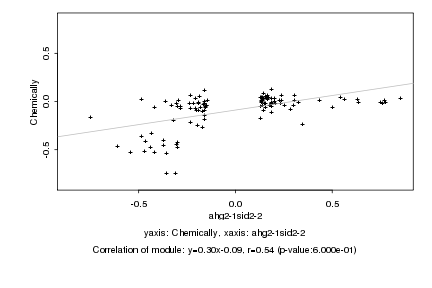
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ahg2-1sid2-2 |
Log2 signal ratio
Chemically |
| 1 |
244901_at |
ATMG00640
|
ORF25 |
0.851 |
0.037 |
| 2 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
0.775 |
-0.004 |
| 3 |
250515_at |
AT5G09570
|
[Cox19-like CHCH family protein] |
0.770 |
0.020 |
| 4 |
244945_at |
ATMG00110
|
ABCI2, ATP-binding cassette I2, CCB206, cytochrome C biogenesis 206 |
0.758 |
-0.011 |
| 5 |
257333_at |
ATMG01360
|
COX1, cytochrome oxidase |
0.747 |
0.000 |
| 6 |
244925_at |
ATMG00510
|
NAD7, NADH dehydrogenase subunit 7 |
0.632 |
-0.001 |
| 7 |
244950_at |
ATMG00160
|
COX2, cytochrome oxidase 2 |
0.626 |
0.028 |
| 8 |
254419_at |
AT4G21490
|
NDB3, NAD(P)H dehydrogenase B3 |
0.559 |
0.021 |
| 9 |
265422_at |
AT2G20800
|
NDB4, NAD(P)H dehydrogenase B4 |
0.539 |
0.042 |
| 10 |
263515_at |
AT2G21640
|
unknown |
0.497 |
-0.057 |
| 11 |
254286_at |
AT4G22950
|
AGL19, AGAMOUS-like 19, GL19 |
0.431 |
0.013 |
| 12 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.345 |
-0.229 |
| 13 |
244921_s_at |
ATMG01000
|
ORF114 |
0.321 |
-0.005 |
| 14 |
256188_at |
AT1G30160
|
unknown |
0.304 |
0.072 |
| 15 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
0.303 |
0.017 |
| 16 |
259481_at |
AT1G18970
|
GLP4, germin-like protein 4 |
0.295 |
-0.037 |
| 17 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.283 |
-0.078 |
| 18 |
257323_at |
ATMG01200
|
ORF294 |
0.249 |
-0.031 |
| 19 |
264868_at |
AT1G24095
|
[Putative thiol-disulphide oxidoreductase DCC] |
0.237 |
0.016 |
| 20 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
0.235 |
0.064 |
| 21 |
252383_at |
AT3G47780
|
ABCA7, ATP-binding cassette A7, ATATH6, A. THALIANA ABC2 HOMOLOG 6, ATH6, ABC2 homolog 6 |
0.231 |
-0.017 |
| 22 |
252864_at |
AT4G39740
|
HCC2, homologue of copper chaperone SCO1 2 |
0.225 |
0.020 |
| 23 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
0.204 |
-0.020 |
| 24 |
262459_at |
AT1G50400
|
[Eukaryotic porin family protein] |
0.201 |
0.033 |
| 25 |
265428_at |
AT2G20720
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.201 |
0.001 |
| 26 |
247612_at |
AT5G60730
|
[Anion-transporting ATPase] |
0.186 |
0.037 |
| 27 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.185 |
0.128 |
| 28 |
265051_at |
AT1G52100
|
[Mannose-binding lectin superfamily protein] |
0.185 |
-0.108 |
| 29 |
257840_at |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
0.184 |
-0.001 |
| 30 |
252281_at |
AT3G49320
|
[Metal-dependent protein hydrolase] |
0.182 |
-0.021 |
| 31 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
0.182 |
-0.051 |
| 32 |
247167_at |
AT5G65850
|
[F-box and associated interaction domains-containing protein] |
0.182 |
-0.014 |
| 33 |
262690_at |
AT1G62720
|
AtNG1, NG1, novel gene 1 |
0.171 |
-0.040 |
| 34 |
245879_at |
AT5G09420
|
AtmtOM64, ATTOC64-V, translocon at the outer membrane of chloroplasts 64-V, MTOM64, ARABIDOPSIS THALIANA TRANSLOCON AT THE OUTER MEMBRANE OF CHLOROPLASTS 64-V, OM64, outer membrane 64, TOC64-V, translocon at the outer membrane of chloroplasts 64-V |
0.170 |
0.049 |
| 35 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
0.165 |
0.066 |
| 36 |
247616_at |
AT5G60260
|
unknown |
0.164 |
0.027 |
| 37 |
247768_at |
AT5G58900
|
[Homeodomain-like transcriptional regulator] |
0.157 |
0.037 |
| 38 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
0.155 |
-0.012 |
| 39 |
258871_at |
AT3G03060
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.152 |
0.062 |
| 40 |
266078_at |
AT2G40670
|
ARR16, response regulator 16, RR16, response regulator 16 |
0.151 |
-0.053 |
| 41 |
264587_at |
AT1G05200
|
ATGLR3.4, glutamate receptor 3.4, GLR3.4, glutamate receptor 3.4, GLUR3 |
0.146 |
-0.016 |
| 42 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
0.145 |
0.088 |
| 43 |
253019_at |
AT4G38010
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.144 |
0.034 |
| 44 |
251811_at |
AT3G54990
|
SMZ, SCHLAFMUTZE |
0.140 |
-0.093 |
| 45 |
249258_at |
AT5G41650
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
0.138 |
0.027 |
| 46 |
256386_at |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
0.137 |
-0.036 |
| 47 |
264895_at |
AT1G23100
|
[GroES-like family protein] |
0.137 |
0.038 |
| 48 |
265061_at |
AT1G61640
|
[Protein kinase superfamily protein] |
0.136 |
0.044 |
| 49 |
248101_at |
AT5G55200
|
MGE1, mitochondrial GrpE 1 |
0.135 |
0.015 |
| 50 |
265964_at |
AT2G37510
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.133 |
0.013 |
| 51 |
250679_at |
AT5G06550
|
JMJ22, Jumonji domain-containing protein 22 |
0.132 |
0.042 |
| 52 |
256319_at |
AT1G35910
|
TPPD, trehalose-6-phosphate phosphatase D |
0.131 |
-0.007 |
| 53 |
251354_at |
AT3G61090
|
[Putative endonuclease or glycosyl hydrolase] |
0.131 |
0.004 |
| 54 |
251915_at |
AT3G53940
|
[Mitochondrial substrate carrier family protein] |
0.128 |
0.045 |
| 55 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.127 |
-0.174 |
| 56 |
247437_at |
AT5G62490
|
ATHVA22B, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE B, HVA22B, HVA22 homologue B |
0.127 |
-0.049 |
| 57 |
253301_at |
AT4G33720
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.751 |
-0.161 |
| 58 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
-0.613 |
-0.465 |
| 59 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.547 |
-0.523 |
| 60 |
262222_at |
AT1G74700
|
NUZ, TRZ1, tRNAse Z1 |
-0.490 |
0.029 |
| 61 |
265102_at |
AT1G30870
|
[Peroxidase superfamily protein] |
-0.487 |
-0.356 |
| 62 |
251226_at |
AT3G62680
|
ATPRP3, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 3, PRP3, proline-rich protein 3 |
-0.474 |
-0.513 |
| 63 |
267457_at |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
-0.467 |
-0.412 |
| 64 |
250778_at |
AT5G05500
|
MOP10 |
-0.441 |
-0.472 |
| 65 |
247477_at |
AT5G62340
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.434 |
-0.322 |
| 66 |
249205_at |
AT5G42600
|
MRN1, marneral synthase 1 |
-0.423 |
-0.060 |
| 67 |
248790_at |
AT5G47450
|
ATTIP2;3, ARABIDOPSIS THALIANA TONOPLAST INTRINSIC PROTEIN 2;3, DELTA-TIP3, DELTA-TONOPLAST INTRINSIC PROTEIN 3, TIP2;3, tonoplast intrinsic protein 2;3 |
-0.422 |
-0.519 |
| 68 |
260758_at |
AT1G48930
|
AtGH9C1, glycosyl hydrolase 9C1, GH9C1, glycosyl hydrolase 9C1 |
-0.374 |
-0.456 |
| 69 |
261099_at |
AT1G62980
|
ATEXP18, ATEXPA18, expansin A18, ATHEXP ALPHA 1.25, EXP18, EXPANSIN 18, EXPA18, expansin A18 |
-0.374 |
-0.402 |
| 70 |
248969_at |
AT5G45310
|
unknown |
-0.365 |
0.008 |
| 71 |
258765_at |
AT3G10710
|
RHS12, root hair specific 12 |
-0.360 |
-0.534 |
| 72 |
249934_at |
AT5G22410
|
RHS18, root hair specific 18 |
-0.358 |
-0.738 |
| 73 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
-0.335 |
-0.032 |
| 74 |
252882_at |
AT4G39675
|
unknown |
-0.322 |
-0.189 |
| 75 |
261562_at |
AT1G01750
|
ADF11, actin depolymerizing factor 11 |
-0.313 |
-0.744 |
| 76 |
253353_at |
AT4G33730
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.309 |
-0.439 |
| 77 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.308 |
-0.016 |
| 78 |
267191_at |
AT2G44110
|
ATMLO15, MILDEW RESISTANCE LOCUS O 15, MLO15, MILDEW RESISTANCE LOCUS O 15 |
-0.304 |
-0.467 |
| 79 |
255814_at |
AT1G19900
|
[glyoxal oxidase-related protein] |
-0.304 |
-0.046 |
| 80 |
259525_at |
AT1G12560
|
ATEXP7, ATEXPA7, expansin A7, ATHEXP ALPHA 1.26, EXP7, EXPA7, expansin A7 |
-0.300 |
-0.420 |
| 81 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
-0.299 |
0.018 |
| 82 |
262004_at |
AT1G64480
|
CBL8, calcineurin B-like protein 8 |
-0.286 |
-0.072 |
| 83 |
267080_at |
AT2G41190
|
[Transmembrane amino acid transporter family protein] |
-0.285 |
-0.046 |
| 84 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
-0.241 |
-0.017 |
| 85 |
247586_at |
AT5G60660
|
PIP2;4, plasma membrane intrinsic protein 2;4, PIP2F |
-0.237 |
-0.213 |
| 86 |
264734_at |
AT1G62280
|
SLAH1, SLAC1 homologue 1 |
-0.234 |
0.068 |
| 87 |
254697_at |
AT4G17970
|
ALMT12, aluminum-activated, malate transporter 12, ATALMT12, QUAC1, quick-activating anion channel 1 |
-0.233 |
-0.069 |
| 88 |
256537_at |
AT1G33340
|
[ENTH/ANTH/VHS superfamily protein] |
-0.219 |
-0.014 |
| 89 |
251842_at |
AT3G54580
|
[Proline-rich extensin-like family protein] |
-0.208 |
-0.068 |
| 90 |
255812_at |
AT4G10310
|
ATHKT1, HKT1, high-affinity K+ transporter 1, HKT1;1 |
-0.207 |
0.033 |
| 91 |
252447_at |
AT3G47040
|
[Glycosyl hydrolase family protein] |
-0.205 |
-0.093 |
| 92 |
250059_at |
AT5G17820
|
[Peroxidase superfamily protein] |
-0.199 |
-0.244 |
| 93 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
-0.196 |
-0.083 |
| 94 |
250138_at |
AT5G14610
|
[DEAD box RNA helicase family protein] |
-0.192 |
-0.001 |
| 95 |
266690_at |
AT2G19900
|
ATNADP-ME1, Arabidopsis thaliana NADP-malic enzyme 1, NADP-ME1, NADP-malic enzyme 1 |
-0.191 |
-0.018 |
| 96 |
262347_at |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
-0.190 |
0.062 |
| 97 |
267343_at |
AT2G44260
|
unknown |
-0.183 |
-0.053 |
| 98 |
254210_at |
AT4G23450
|
AIRP1, ABA Insensitive RING Protein 1, AtAIRP1 |
-0.174 |
-0.097 |
| 99 |
247536_at |
AT5G61650
|
CYCP4, CYCP4;2, CYCLIN P4;2 |
-0.174 |
-0.265 |
| 100 |
245674_at |
AT1G56680
|
[Chitinase family protein] |
-0.169 |
-0.025 |
| 101 |
266196_at |
AT2G39110
|
[Protein kinase superfamily protein] |
-0.165 |
-0.137 |
| 102 |
249860_at |
AT5G22860
|
[Serine carboxypeptidase S28 family protein] |
-0.165 |
0.122 |
| 103 |
252209_at |
AT3G50400
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.164 |
0.003 |
| 104 |
249522_at |
AT5G38700
|
unknown |
-0.164 |
-0.089 |
| 105 |
256283_at |
AT3G12540
|
unknown |
-0.161 |
-0.186 |
| 106 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
-0.160 |
-0.020 |
| 107 |
250860_at |
AT5G04770
|
ATCAT6, ARABIDOPSIS THALIANA CATIONIC AMINO ACID TRANSPORTER 6, CAT6, cationic amino acid transporter 6 |
-0.159 |
-0.047 |
| 108 |
246420_at |
AT5G16870
|
[Peptidyl-tRNA hydrolase II (PTH2) family protein] |
-0.157 |
-0.062 |
| 109 |
264144_at |
AT1G79320
|
AtMC6, metacaspase 6, AtMCP2c, metacaspase 2c, MC6, metacaspase 6, MCP2c, metacaspase 2c |
-0.154 |
-0.034 |
| 110 |
258905_at |
AT3G06390
|
[Uncharacterised protein family (UPF0497)] |
-0.150 |
-0.038 |
| 111 |
253993_at |
AT4G26070
|
ATMEK1, MEK1, MAP kinase/ ERK kinase 1, MKK1, MITOGEN ACTIVATED PROTEIN KINASE KINASE 1, NMAPKK |
-0.149 |
0.014 |