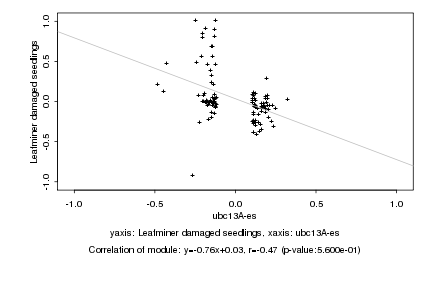
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ubc13A-es |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
260813_at |
AT1G43700
|
SUE3, sulphate utilization efficiency 3, VIP1, VIRE2-interacting protein 1 |
0.323 |
0.036 |
| 2 |
244998_at |
ATCG00180
|
RPOC1 |
0.244 |
-0.075 |
| 3 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
0.235 |
-0.307 |
| 4 |
245047_at |
ATCG00020
|
PSBA, photosystem II reaction center protein A |
0.225 |
-0.040 |
| 5 |
244937_at |
ATCG01110
|
NDHH, NAD(P)H dehydrogenase subunit H |
0.222 |
-0.243 |
| 6 |
267085_at |
AT2G32620
|
ATCSLB02, cellulose synthase-like B, ATCSLB2, CELLULOSE SYNTHASE LIKE B2, CSLB02, CELLULOSE SYNTHASE LIKE B2, CSLB02, cellulose synthase-like B |
0.209 |
-0.048 |
| 7 |
255957_at |
AT1G22160
|
unknown |
0.202 |
-0.196 |
| 8 |
244995_at |
ATCG00150
|
ATPI |
0.199 |
-0.089 |
| 9 |
266611_at |
AT2G14960
|
GH3.1 |
0.198 |
0.043 |
| 10 |
253182_at |
AT4G35190
|
LOG5, LONELY GUY 5 |
0.197 |
0.086 |
| 11 |
244961_at |
ATCG01040
|
YCF5 |
0.196 |
-0.045 |
| 12 |
255252_at |
AT4G04990
|
unknown |
0.188 |
-0.011 |
| 13 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
0.187 |
0.293 |
| 14 |
245024_at |
ATCG00120
|
ATPA, ATP synthase subunit alpha |
0.184 |
-0.131 |
| 15 |
244903_at |
ATMG00660
|
ORF149 |
0.183 |
0.075 |
| 16 |
244999_at |
ATCG00190
|
RPOB, RNA polymerase subunit beta |
0.181 |
-0.075 |
| 17 |
251197_at |
AT3G62960
|
[Thioredoxin superfamily protein] |
0.181 |
0.043 |
| 18 |
257274_at |
AT3G14510
|
[Polyprenyl synthetase family protein] |
0.177 |
-0.050 |
| 19 |
260007_at |
AT1G67870
|
[glycine-rich protein] |
0.173 |
-0.048 |
| 20 |
244971_at |
ATCG00670
|
CLPP1, CASEINOLYTIC PROTEASE P 1, PCLPP, plastid-encoded CLP P |
0.172 |
-0.018 |
| 21 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
0.164 |
-0.071 |
| 22 |
245006_at |
ATCG00340
|
PSAB |
0.161 |
-0.022 |
| 23 |
264598_at |
AT1G04610
|
YUC3, YUCCA 3 |
0.160 |
-0.086 |
| 24 |
245026_at |
ATCG00140
|
ATPH |
0.158 |
-0.061 |
| 25 |
265267_at |
AT2G42920
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.156 |
-0.118 |
| 26 |
253495_at |
AT4G31850
|
PGR3, proton gradient regulation 3 |
0.156 |
-0.346 |
| 27 |
245947_at |
AT5G19530
|
ACL5, ACAULIS 5 |
0.154 |
-0.276 |
| 28 |
250828_at |
AT5G05250
|
unknown |
0.149 |
-0.367 |
| 29 |
267197_at |
AT2G30960
|
unknown |
0.140 |
-0.255 |
| 30 |
246891_at |
AT5G25490
|
[Ran BP2/NZF zinc finger-like superfamily protein] |
0.139 |
-0.159 |
| 31 |
250262_at |
AT5G13410
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
0.134 |
-0.085 |
| 32 |
263909_at |
AT2G36490
|
AtROS1, DML1, demeter-like 1, ROS1, REPRESSOR OF SILENCING1 |
0.130 |
-0.404 |
| 33 |
249986_at |
AT5G18460
|
unknown |
0.122 |
-0.290 |
| 34 |
247239_at |
AT5G64640
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.122 |
-0.039 |
| 35 |
245499_at |
AT4G16480
|
ATINT4, INT4, inositol transporter 4 |
0.121 |
0.109 |
| 36 |
244913_at |
ATMG00840
|
ORF121B |
0.121 |
0.025 |
| 37 |
257541_at |
AT3G25950
|
[TRAM, LAG1 and CLN8 (TLC) lipid-sensing domain containing protein] |
0.121 |
-0.073 |
| 38 |
254034_at |
AT4G25960
|
ABCB2, ATP-binding cassette B2, PGP2, P-glycoprotein 2 |
0.119 |
-0.236 |
| 39 |
253158_at |
AT4G35780
|
STY17, serine/threonine/tyrosine kinase 17 |
0.119 |
-0.254 |
| 40 |
264481_at |
AT1G77200
|
[Integrase-type DNA-binding superfamily protein] |
0.118 |
0.050 |
| 41 |
263164_at |
AT1G03070
|
AtLFG4, LFG4, LIFEGUARD 4 |
0.118 |
-0.035 |
| 42 |
263043_at |
AT1G23350
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.115 |
-0.060 |
| 43 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
0.114 |
-0.050 |
| 44 |
251940_at |
AT3G53450
|
LOG4, LONELY GUY 4 |
0.112 |
0.078 |
| 45 |
258603_at |
AT3G02990
|
ATHSFA1E, heat shock transcription factor A1E, HSFA1E, heat shock transcription factor A1E |
0.112 |
0.122 |
| 46 |
257421_at |
AT1G12030
|
unknown |
0.111 |
-0.375 |
| 47 |
264573_at |
AT1G05310
|
[Pectin lyase-like superfamily protein] |
0.109 |
-0.150 |
| 48 |
261003_at |
AT1G26500
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.109 |
-0.125 |
| 49 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
0.108 |
-0.236 |
| 50 |
262969_at |
AT1G75710
|
[C2H2-like zinc finger protein] |
0.108 |
-0.384 |
| 51 |
246792_at |
AT5G27290
|
unknown |
0.108 |
-0.237 |
| 52 |
258299_at |
AT3G23410
|
ATFAO3, ARABIDOPSIS FATTY ALCOHOL OXIDASE 3, FAO3, fatty alcohol oxidase 3 |
0.106 |
-0.268 |
| 53 |
246329_at |
AT3G43610
|
[Spc97 / Spc98 family of spindle pole body (SBP) component] |
0.104 |
-0.245 |
| 54 |
255374_at |
AT4G03770
|
[gypsy-like retrotransposon family, has a 2.8e-206 P-value blast match to GB:AAD11615 prpol (gypsy_Ty3-element) (Zea mays)] |
0.104 |
-0.006 |
| 55 |
245486_x_at |
AT4G16240
|
unknown |
0.103 |
0.099 |
| 56 |
260486_at |
AT1G51550
|
[Kelch repeat-containing F-box family protein] |
0.103 |
0.043 |
| 57 |
261331_at |
AT1G44935
|
SADHU9-1, sadhu non-coding retrotransposon 9-1 |
0.103 |
-0.012 |
| 58 |
248350_at |
AT5G52160
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.101 |
0.019 |
| 59 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.488 |
0.224 |
| 60 |
255883_at |
AT1G20270
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.451 |
0.131 |
| 61 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
-0.432 |
0.477 |
| 62 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.267 |
-0.918 |
| 63 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
-0.254 |
1.022 |
| 64 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
-0.242 |
0.489 |
| 65 |
259251_at |
AT3G07600
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.234 |
0.081 |
| 66 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
-0.224 |
-0.255 |
| 67 |
265796_at |
AT2G35730
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.214 |
0.573 |
| 68 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
-0.208 |
0.806 |
| 69 |
257720_at |
AT3G18450
|
[PLAC8 family protein] |
-0.207 |
0.010 |
| 70 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
-0.205 |
0.856 |
| 71 |
254743_at |
AT4G13420
|
ATHAK5, ARABIDOPSIS THALIANA HIGH AFFINITY K+ TRANSPORTER 5, HAK5, high affinity K+ transporter 5 |
-0.201 |
0.007 |
| 72 |
246912_at |
AT5G25820
|
[Exostosin family protein] |
-0.201 |
0.082 |
| 73 |
250702_at |
AT5G06730
|
[Peroxidase superfamily protein] |
-0.194 |
0.111 |
| 74 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
-0.189 |
0.915 |
| 75 |
248130_at |
AT5G54790
|
unknown |
-0.188 |
-0.004 |
| 76 |
249108_at |
AT5G43690
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.185 |
0.016 |
| 77 |
247899_at |
AT5G57345
|
unknown |
-0.184 |
-0.002 |
| 78 |
260051_at |
AT1G78210
|
[alpha/beta-Hydrolases superfamily protein] |
-0.178 |
0.472 |
| 79 |
251405_at |
AT3G60330
|
AHA7, H(+)-ATPase 7, HA7, H(+)-ATPase 7 |
-0.177 |
-0.041 |
| 80 |
255360_at |
AT4G03960
|
AtPFA-DSP4, PFA-DSP4, plant and fungi atypical dual-specificity phosphatase 4 |
-0.177 |
0.007 |
| 81 |
264014_at |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
-0.173 |
-0.217 |
| 82 |
254912_at |
AT4G11230
|
[Riboflavin synthase-like superfamily protein] |
-0.169 |
-0.027 |
| 83 |
252510_at |
AT3G46270
|
[receptor protein kinase-related] |
-0.165 |
-0.003 |
| 84 |
247314_at |
AT5G64000
|
ATSAL2, SAL2 |
-0.159 |
0.016 |
| 85 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
-0.159 |
0.395 |
| 86 |
255046_at |
AT4G09650
|
ATPD, ATP synthase delta-subunit gene, PDE332, PIGMENT DEFECTIVE 332 |
-0.153 |
-0.125 |
| 87 |
254914_at |
AT4G11290
|
[Peroxidase superfamily protein] |
-0.152 |
-0.010 |
| 88 |
265279_at |
AT2G28460
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.152 |
-0.188 |
| 89 |
251884_at |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.152 |
0.693 |
| 90 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
-0.150 |
0.329 |
| 91 |
252859_at |
AT4G39780
|
[Integrase-type DNA-binding superfamily protein] |
-0.149 |
0.242 |
| 92 |
248537_at |
AT5G50100
|
[Putative thiol-disulphide oxidoreductase DCC] |
-0.147 |
-0.038 |
| 93 |
248931_at |
AT5G46040
|
[Major facilitator superfamily protein] |
-0.147 |
0.060 |
| 94 |
258477_at |
AT3G02680
|
ATNBS1, NBS1, nijmegen breakage syndrome 1 |
-0.143 |
-0.042 |
| 95 |
250323_at |
AT5G12880
|
[proline-rich family protein] |
-0.143 |
0.571 |
| 96 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
-0.143 |
0.687 |
| 97 |
256788_at |
AT3G13730
|
CYP90D1, cytochrome P450, family 90, subfamily D, polypeptide 1 |
-0.140 |
0.213 |
| 98 |
267526_at |
AT2G30570
|
PSBW, photosystem II reaction center W |
-0.139 |
0.001 |
| 99 |
262748_at |
AT1G28610
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.137 |
-0.015 |
| 100 |
249773_at |
AT5G24140
|
SQP2, squalene monooxygenase 2 |
-0.135 |
0.009 |
| 101 |
266123_at |
AT2G45180
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.134 |
-0.149 |
| 102 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
-0.134 |
0.903 |
| 103 |
247670_at |
AT5G60190
|
[Cysteine proteinases superfamily protein] |
-0.134 |
0.090 |
| 104 |
250603_at |
AT5G07820
|
[Plant calmodulin-binding protein-related] |
-0.134 |
0.031 |
| 105 |
252395_at |
AT3G47950
|
AHA4, H(+)-ATPase 4, HA4, H(+)-ATPase 4 |
-0.133 |
0.043 |
| 106 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
-0.130 |
0.813 |
| 107 |
245019_at |
ATCG00530
|
YCF10 |
-0.129 |
-0.066 |
| 108 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.129 |
1.015 |
| 109 |
252403_at |
AT3G48080
|
[alpha/beta-Hydrolases superfamily protein] |
-0.128 |
0.044 |
| 110 |
250683_x_at |
AT5G06640
|
EXT10, extensin 10 |
-0.128 |
-0.064 |
| 111 |
262305_at |
AT1G70950
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.126 |
-0.053 |
| 112 |
253323_at |
AT4G33920
|
APD5, Arabidopsis Pp2c clade D 5 |
-0.126 |
0.464 |
| 113 |
258342_at |
AT3G22800
|
[Leucine-rich repeat (LRR) family protein] |
-0.126 |
-0.017 |
| 114 |
260060_at |
AT1G73680
|
ALPHA DOX2, alpha dioxygenase, alpha-DOX2, alpha-dioxygenase 2 |
-0.123 |
0.052 |
| 115 |
247998_at |
AT5G56200
|
[C2H2 type zinc finger transcription factor family] |
-0.121 |
-0.032 |