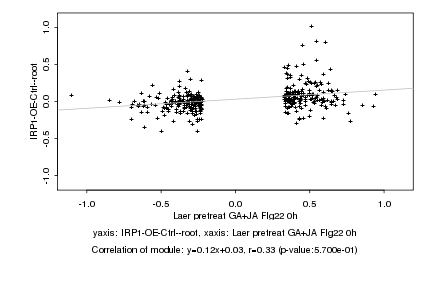
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Laer pretreat GA+JA Flg22 0h |
Log2 signal ratio
IRP1-OE-Ctrl--root |
| 1 |
253060_at |
AT4G37710
|
[VQ motif-containing protein] |
0.941 |
0.100 |
| 2 |
266232_at |
AT2G02310
|
AtPP2-B6, phloem protein 2-B6, PP2-B6, phloem protein 2-B6 |
0.926 |
-0.065 |
| 3 |
259036_at |
AT3G09220
|
LAC7, laccase 7 |
0.853 |
-0.048 |
| 4 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
0.774 |
-0.263 |
| 5 |
260492_at |
AT2G41850
|
ADPG2, ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 2, PGAZAT, polygalacturonase abscission zone A. thaliana |
0.755 |
-0.149 |
| 6 |
256762_at |
AT3G25655
|
IDL1, inflorescence deficient in abscission (IDA)-like 1 |
0.737 |
0.103 |
| 7 |
249559_at |
AT5G38320
|
unknown |
0.724 |
0.016 |
| 8 |
258182_at |
AT3G21500
|
DXL1, DXS-like 1, DXPS1, 1-deoxy-D-xylulose 5-phosphate synthase 1, DXS2, 1-deoxy-D-xylulose 5-phosphate (DXP) synthase 2 |
0.721 |
-0.038 |
| 9 |
266455_at |
AT2G22760
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.682 |
0.034 |
| 10 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
0.681 |
0.149 |
| 11 |
260802_at |
AT1G78400
|
[Pectin lyase-like superfamily protein] |
0.674 |
0.063 |
| 12 |
252212_at |
AT3G50310
|
MAPKKK20, mitogen-activated protein kinase kinase kinase 20, MKKK20, MAPKK kinase 20 |
0.669 |
-0.051 |
| 13 |
256937_at |
AT3G22620
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.666 |
0.082 |
| 14 |
261278_at |
AT1G05800
|
DGL, DONGLE |
0.658 |
0.011 |
| 15 |
251281_at |
AT3G61640
|
AGP20, arabinogalactan protein 20, AtAGP20, Arabinogalactan protein 20 |
0.649 |
0.143 |
| 16 |
249154_at |
AT5G43410
|
[Integrase-type DNA-binding superfamily protein] |
0.646 |
0.010 |
| 17 |
248227_at |
AT5G53820
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.643 |
0.144 |
| 18 |
264886_at |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
0.641 |
0.149 |
| 19 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
0.641 |
-0.027 |
| 20 |
246978_at |
AT5G24910
|
CYP714A1, cytochrome P450, family 714, subfamily A, polypeptide 1, ELA1, EUI-like p450 A1 |
0.640 |
-0.004 |
| 21 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
0.639 |
0.446 |
| 22 |
267276_at |
AT2G30130
|
ASL5, LBD12, PCK1, PEACOCK 1 |
0.626 |
0.155 |
| 23 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
0.624 |
0.255 |
| 24 |
247253_at |
AT5G64790
|
[O-Glycosyl hydrolases family 17 protein] |
0.614 |
0.025 |
| 25 |
253322_at |
AT4G33980
|
unknown |
0.611 |
-0.089 |
| 26 |
253743_at |
AT4G28940
|
[Phosphorylase superfamily protein] |
0.604 |
-0.057 |
| 27 |
253259_at |
AT4G34410
|
RRTF1, redox responsive transcription factor 1 |
0.600 |
0.807 |
| 28 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.593 |
0.001 |
| 29 |
261814_at |
AT1G08310
|
[alpha/beta-Hydrolases superfamily protein] |
0.592 |
-0.219 |
| 30 |
266273_at |
AT2G29410
|
ATMTPB1, MTPB1, metal tolerance protein B1 |
0.592 |
0.051 |
| 31 |
264425_at |
AT1G61750
|
[Receptor-like protein kinase-related family protein] |
0.591 |
0.014 |
| 32 |
252200_at |
AT3G50280
|
[HXXXD-type acyl-transferase family protein] |
0.590 |
0.369 |
| 33 |
264543_at |
AT1G55790
|
[FUNCTIONS IN: metal ion binding] |
0.589 |
0.127 |
| 34 |
252638_at |
AT3G44540
|
FAR4, fatty acid reductase 4 |
0.585 |
-0.010 |
| 35 |
245076_at |
AT2G23170
|
GH3.3 |
0.578 |
0.237 |
| 36 |
255811_at |
AT4G10250
|
ATHSP22.0 |
0.573 |
-0.046 |
| 37 |
247104_at |
AT5G66620
|
DAR6, DA1-related protein 6 |
0.573 |
0.033 |
| 38 |
266271_at |
AT2G29440
|
ATGSTU6, glutathione S-transferase tau 6, GST24, GLUTATHIONE S-TRANSFERASE 24, GSTU6, glutathione S-transferase tau 6 |
0.571 |
0.258 |
| 39 |
257564_at |
AT3G28610
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.564 |
0.026 |
| 40 |
245034_at |
AT2G26390
|
[Serine protease inhibitor (SERPIN) family protein] |
0.561 |
0.150 |
| 41 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.552 |
0.085 |
| 42 |
251114_at |
AT5G01380
|
[Homeodomain-like superfamily protein] |
0.551 |
0.017 |
| 43 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.550 |
0.222 |
| 44 |
259866_at |
AT1G76640
|
CML39, CALMODULIN LIKE 39 |
0.543 |
0.567 |
| 45 |
259442_at |
AT1G02310
|
MAN1, endo-beta-mannanase 1 |
0.542 |
0.203 |
| 46 |
266132_at |
AT2G45130
|
ATSPX3, ARABIDOPSIS THALIANA SPX DOMAIN GENE 3, SPX3, SPX domain gene 3 |
0.542 |
0.062 |
| 47 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.540 |
0.824 |
| 48 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.535 |
0.050 |
| 49 |
259400_at |
AT1G17750
|
AtPEPR2, PEP1 RECEPTOR 2, PEPR2, PEP1 receptor 2 |
0.532 |
0.260 |
| 50 |
245447_at |
AT4G16820
|
DALL1, DAD1-Like Lipase 1, PLA-I{beta]2, phospholipase A I beta 2 |
0.531 |
0.001 |
| 51 |
259947_at |
AT1G71530
|
[Protein kinase superfamily protein] |
0.528 |
-0.020 |
| 52 |
247913_at |
AT5G57510
|
unknown |
0.527 |
0.256 |
| 53 |
263916_at |
AT2G36440
|
unknown |
0.521 |
-0.011 |
| 54 |
266486_at |
AT2G47950
|
unknown |
0.519 |
0.098 |
| 55 |
265658_at |
AT2G13810
|
ALD1, AGD2-like defense response protein 1, AtALD1, EDTS5, eds two suppressor 5 |
0.517 |
0.102 |
| 56 |
251971_at |
AT3G53160
|
UGT73C7, UDP-glucosyl transferase 73C7 |
0.514 |
0.091 |
| 57 |
262649_at |
AT1G14040
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.513 |
0.052 |
| 58 |
251846_at |
AT3G54560
|
HTA11, histone H2A 11 |
0.510 |
0.037 |
| 59 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
0.510 |
0.248 |
| 60 |
253832_at |
AT4G27654
|
unknown |
0.509 |
1.025 |
| 61 |
258796_at |
AT3G04630
|
WDL1, WVD2-like 1 |
0.505 |
-0.108 |
| 62 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.504 |
0.269 |
| 63 |
259913_at |
AT1G72660
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.503 |
0.028 |
| 64 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
0.498 |
0.150 |
| 65 |
248333_at |
AT5G52390
|
[PAR1 protein] |
0.497 |
0.083 |
| 66 |
261545_at |
AT1G63530
|
unknown |
0.496 |
-0.190 |
| 67 |
264646_at |
AT1G08860
|
BON3, BONZAI 3 |
0.494 |
0.113 |
| 68 |
265723_at |
AT2G32140
|
[transmembrane receptors] |
0.485 |
0.275 |
| 69 |
259251_at |
AT3G07600
|
[Heavy metal transport/detoxification superfamily protein ] |
0.481 |
0.318 |
| 70 |
262307_at |
AT1G71000
|
[Chaperone DnaJ-domain superfamily protein] |
0.480 |
0.088 |
| 71 |
249495_at |
AT5G39100
|
GLP6, germin-like protein 6 |
0.472 |
0.231 |
| 72 |
251207_at |
AT3G63050
|
unknown |
0.471 |
-0.042 |
| 73 |
253884_at |
AT4G27670
|
HSP21, heat shock protein 21 |
0.468 |
0.028 |
| 74 |
259282_at |
AT3G11430
|
ATGPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5, GPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5 |
0.468 |
0.010 |
| 75 |
247789_at |
AT5G58680
|
[ARM repeat superfamily protein] |
0.467 |
0.256 |
| 76 |
245440_at |
AT4G16680
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.466 |
0.065 |
| 77 |
256109_at |
AT1G16950
|
unknown |
0.457 |
0.109 |
| 78 |
256009_at |
AT1G19210
|
[Integrase-type DNA-binding superfamily protein] |
0.457 |
0.511 |
| 79 |
251706_at |
AT3G56620
|
UMAMIT10, Usually multiple acids move in and out Transporters 10 |
0.455 |
-0.224 |
| 80 |
247333_at |
AT5G63600
|
ATFLS5, FLS5, flavonol synthase 5 |
0.453 |
0.091 |
| 81 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
0.447 |
0.767 |
| 82 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
0.445 |
0.175 |
| 83 |
254062_at |
AT4G25380
|
AtSAP10, Arabidopsis thaliana stress-associated protein 10, SAP10, stress-associated protein 10 |
0.444 |
0.077 |
| 84 |
254323_at |
AT4G22620
|
SAUR34, SMALL AUXIN UPREGULATED RNA 34 |
0.443 |
0.087 |
| 85 |
257840_at |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
0.443 |
0.357 |
| 86 |
265732_at |
AT2G01300
|
unknown |
0.441 |
0.148 |
| 87 |
263754_at |
AT2G21510
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.439 |
-0.092 |
| 88 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
0.438 |
0.117 |
| 89 |
248618_at |
AT5G49620
|
AtMYB78, myb domain protein 78, MYB78, myb domain protein 78 |
0.436 |
0.023 |
| 90 |
263684_at |
AT1G26900
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.435 |
-0.014 |
| 91 |
258377_at |
AT3G17690
|
ATCNGC19, CYCLIC NUCLEOTIDE-GATED CHANNEL 19, CNGC19, cyclic nucleotide gated channel 19 |
0.434 |
0.037 |
| 92 |
261109_at |
AT1G75450
|
ATCKX5, ARABIDOPSIS THALIANA CYTOKININ OXIDASE 5, ATCKX6, CYTOKININ OXIDASE 6, CKX5, cytokinin oxidase 5 |
0.434 |
0.036 |
| 93 |
252340_at |
AT3G48920
|
AtMYB45, myb domain protein 45, MYB45, myb domain protein 45 |
0.432 |
0.049 |
| 94 |
256285_at |
AT3G12510
|
[MADS-box family protein] |
0.430 |
0.010 |
| 95 |
264832_at |
AT1G03660
|
[Ankyrin-repeat containing protein] |
0.428 |
-0.239 |
| 96 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.428 |
0.112 |
| 97 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.426 |
-0.064 |
| 98 |
247208_at |
AT5G64870
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
0.425 |
0.305 |
| 99 |
260668_at |
AT1G19530
|
unknown |
0.425 |
-0.217 |
| 100 |
258338_at |
AT3G16150
|
ASPGB1, asparaginase B1 |
0.423 |
0.138 |
| 101 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
0.420 |
0.141 |
| 102 |
264469_at |
AT1G67100
|
LBD40, LOB domain-containing protein 40 |
0.413 |
0.096 |
| 103 |
258516_at |
AT3G06490
|
AtMYB108, myb domain protein 108, BOS1, BOTRYTIS-SUSCEPTIBLE1, MYB108, myb domain protein 108 |
0.413 |
0.044 |
| 104 |
249705_at |
AT5G35580
|
[Protein kinase superfamily protein] |
0.412 |
0.123 |
| 105 |
249971_at |
AT5G19110
|
[Eukaryotic aspartyl protease family protein] |
0.410 |
0.475 |
| 106 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
0.408 |
-0.295 |
| 107 |
253325_at |
AT4G33925
|
SSN2, suppressor of sni1 2 |
0.407 |
-0.134 |
| 108 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
0.402 |
0.052 |
| 109 |
259327_at |
AT3G16460
|
JAL34, jacalin-related lectin 34 |
0.401 |
0.100 |
| 110 |
259374_at |
AT3G16280
|
[Integrase-type DNA-binding superfamily protein] |
0.400 |
-0.075 |
| 111 |
257226_at |
AT3G27880
|
unknown |
0.399 |
0.143 |
| 112 |
260917_at |
AT1G02700
|
unknown |
0.399 |
0.062 |
| 113 |
247360_at |
AT5G63450
|
CYP94B1, cytochrome P450, family 94, subfamily B, polypeptide 1 |
0.394 |
0.094 |
| 114 |
259150_at |
AT3G10320
|
[Glycosyltransferase family 61 protein] |
0.394 |
0.144 |
| 115 |
245243_at |
AT1G44414
|
unknown |
0.394 |
-0.007 |
| 116 |
258760_at |
AT3G10780
|
[emp24/gp25L/p24 family/GOLD family protein] |
0.394 |
0.030 |
| 117 |
261069_at |
AT1G07410
|
ATRAB-A2B, ARABIDOPSIS RAB GTPASE HOMOLOG A2B, ATRABA2B, RAB GTPase homolog A2B, RAB-A2B, RAB GTPASE HOMOLOG A2B, RABA2b, RAB GTPase homolog A2B |
0.393 |
-0.010 |
| 118 |
247427_at |
AT5G62580
|
[ARM repeat superfamily protein] |
0.392 |
-0.014 |
| 119 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.389 |
0.082 |
| 120 |
250246_at |
AT5G13700
|
APAO, ATPAO1, polyamine oxidase 1, PAO1, polyamine oxidase 1 |
0.389 |
-0.013 |
| 121 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
0.388 |
0.055 |
| 122 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
0.388 |
0.218 |
| 123 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
0.387 |
0.037 |
| 124 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
0.385 |
-0.025 |
| 125 |
259595_at |
AT1G28050
|
BBX13, B-box domain protein 13 |
0.385 |
0.096 |
| 126 |
254630_at |
AT4G18360
|
GOX3, glycolate oxidase 3 |
0.384 |
0.108 |
| 127 |
266756_at |
AT2G46950
|
CYP709B2, cytochrome P450, family 709, subfamily B, polypeptide 2 |
0.383 |
-0.028 |
| 128 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.383 |
0.005 |
| 129 |
257824_at |
AT3G25290
|
[Auxin-responsive family protein] |
0.382 |
0.016 |
| 130 |
261838_at |
AT1G16030
|
Hsp70b, heat shock protein 70B |
0.382 |
-0.025 |
| 131 |
252230_at |
AT3G49810
|
[ARM repeat superfamily protein] |
0.382 |
-0.035 |
| 132 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
0.380 |
0.126 |
| 133 |
260204_at |
AT1G52900
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.380 |
-0.040 |
| 134 |
262813_at |
AT1G11670
|
[MATE efflux family protein] |
0.375 |
0.029 |
| 135 |
264053_at |
AT2G22560
|
NET2D, Networked 2D |
0.371 |
0.016 |
| 136 |
255536_at |
AT4G01575
|
[serine protease inhibitor, Kazal-type family protein] |
0.371 |
-0.058 |
| 137 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
0.370 |
0.021 |
| 138 |
262061_at |
AT1G80110
|
ATPP2-B11, phloem protein 2-B11, PP2-B11, phloem protein 2-B11 |
0.369 |
-0.021 |
| 139 |
265216_at |
AT1G05100
|
MAPKKK18, mitogen-activated protein kinase kinase kinase 18 |
0.368 |
0.183 |
| 140 |
266447_at |
AT2G43290
|
MSS3, multicopy suppressors of snf4 deficiency in yeast 3 |
0.366 |
0.123 |
| 141 |
253100_at |
AT4G37400
|
CYP81F3, cytochrome P450, family 81, subfamily F, polypeptide 3 |
0.366 |
0.365 |
| 142 |
254201_at |
AT4G24130
|
unknown |
0.366 |
0.100 |
| 143 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.366 |
0.091 |
| 144 |
255950_at |
AT1G22110
|
[structural constituent of ribosome] |
0.366 |
0.087 |
| 145 |
257644_at |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
0.364 |
0.330 |
| 146 |
264978_at |
AT1G27120
|
[Galactosyltransferase family protein] |
0.361 |
-0.076 |
| 147 |
250024_at |
AT5G18270
|
ANAC087, Arabidopsis NAC domain containing protein 87 |
0.360 |
-0.007 |
| 148 |
263164_at |
AT1G03070
|
AtLFG4, LFG4, LIFEGUARD 4 |
0.360 |
0.014 |
| 149 |
260885_at |
AT1G29230
|
ATCIPK18, ATWL1, WPL4-LIKE 1, CIPK18, CBL-interacting protein kinase 18, SnRK3.20, SNF1-RELATED PROTEIN KINASE 3.20, WL1, WPL4-LIKE 1 |
0.359 |
0.053 |
| 150 |
260506_at |
AT1G47210
|
CYCA3;2, cyclin-dependent protein kinase 3;2 |
0.358 |
0.119 |
| 151 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.357 |
0.109 |
| 152 |
262755_at |
AT1G16360
|
[LEM3 (ligand-effect modulator 3) family protein / CDC50 family protein] |
0.357 |
0.031 |
| 153 |
261344_at |
AT1G79710
|
[Major facilitator superfamily protein] |
0.357 |
0.047 |
| 154 |
253799_at |
AT4G28140
|
[Integrase-type DNA-binding superfamily protein] |
0.357 |
0.023 |
| 155 |
258953_at |
AT3G01430
|
unknown |
0.355 |
0.022 |
| 156 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.355 |
0.100 |
| 157 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
0.354 |
0.498 |
| 158 |
247691_at |
AT5G59720
|
HSP18.2, heat shock protein 18.2 |
0.352 |
-0.156 |
| 159 |
259780_at |
AT1G29630
|
[5'-3' exonuclease family protein] |
0.352 |
0.040 |
| 160 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
0.351 |
-0.069 |
| 161 |
248154_at |
AT5G54400
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.351 |
0.070 |
| 162 |
261700_at |
AT1G32690
|
unknown |
0.351 |
0.113 |
| 163 |
245112_at |
AT2G41540
|
GPDHC1 |
0.349 |
0.006 |
| 164 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.349 |
0.370 |
| 165 |
253632_at |
AT4G30430
|
TET9, tetraspanin9 |
0.346 |
0.099 |
| 166 |
263734_at |
AT1G60030
|
ATNAT7, ARABIDOPSIS NUCLEOBASE-ASCORBATE TRANSPORTER 7, NAT7, nucleobase-ascorbate transporter 7 |
0.345 |
-0.149 |
| 167 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
0.344 |
0.324 |
| 168 |
260365_at |
AT1G70630
|
RAY1, REDUCED ARABINOSE YARIV 1 |
0.344 |
-0.081 |
| 169 |
256430_at |
AT3G11020
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2B, DRE/CRT-binding protein 2B |
0.344 |
0.109 |
| 170 |
254069_at |
AT4G25434
|
ATNUDT10, nudix hydrolase homolog 10, NUDT10, nudix hydrolase homolog 10 |
0.344 |
0.191 |
| 171 |
248316_at |
AT5G52670
|
[Copper transport protein family] |
0.344 |
0.449 |
| 172 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
0.343 |
0.138 |
| 173 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
0.343 |
0.185 |
| 174 |
263073_at |
AT2G17500
|
[Auxin efflux carrier family protein] |
0.340 |
0.032 |
| 175 |
264518_at |
AT1G09980
|
[Putative serine esterase family protein] |
0.338 |
0.391 |
| 176 |
252006_at |
AT3G52820
|
ATPAP22, PURPLE ACID PHOSPHATASE 22, PAP22, purple acid phosphatase 22 |
0.336 |
-0.145 |
| 177 |
259418_at |
AT1G02390
|
ATGPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2, GPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2 |
0.336 |
-0.057 |
| 178 |
247290_at |
AT5G64450
|
unknown |
0.335 |
-0.021 |
| 179 |
260023_at |
AT1G30040
|
ATGA2OX2, gibberellin 2-oxidase, GA2OX2, gibberellin 2-oxidase, GA2OX2, GIBBERELLIN 2-OXIDASE 2 |
0.334 |
0.039 |
| 180 |
267280_at |
AT2G19450
|
ABX45, AS11, ATDGAT, Arabidopsis thaliana acyl-CoA:diacylglycerol acyltransferase, AtDGAT1, DGAT1, acyl-CoA:diacylglycerol acyltransferase 1, RDS1, TAG1, TRIACYLGLYCEROL BIOSYNTHESIS DEFECT 1 |
0.332 |
0.004 |
| 181 |
252179_at |
AT3G50760
|
GATL2, galacturonosyltransferase-like 2 |
0.332 |
0.087 |
| 182 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.331 |
0.068 |
| 183 |
246584_at |
AT5G14730
|
unknown |
0.330 |
0.463 |
| 184 |
248410_at |
AT5G51570
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
0.330 |
0.035 |
| 185 |
260712_at |
AT1G17550
|
AtHAB2, HAB2, homology to ABI2 |
0.328 |
0.055 |
| 186 |
257271_at |
AT3G28007
|
AtSWEET4, SWEET4 |
0.328 |
0.072 |
| 187 |
246310_at |
AT3G51895
|
AST12, SULTR3;1, sulfate transporter 3;1 |
0.328 |
0.087 |
| 188 |
266421_at |
AT2G38540
|
ATLTP1, ARABIDOPSIS THALIANA LIPID TRANSFER PROTEIN 1, LP1, lipid transfer protein 1, LTP1, LIPID TRANSFER PROTEIN 1 |
-1.108 |
0.084 |
| 189 |
263709_at |
AT1G09310
|
unknown |
-0.854 |
0.027 |
| 190 |
259892_at |
AT1G72610
|
ATGER1, A. THALIANA GERMIN-LIKE PROTEIN 1, GER1, germin-like protein 1, GLP1, GERMIN-LIKE PROTEIN 1 |
-0.785 |
-0.009 |
| 191 |
262281_at |
AT1G68570
|
AtNPF3.1, NPF3.1, NRT1/ PTR family 3.1 |
-0.706 |
-0.235 |
| 192 |
260267_at |
AT1G68530
|
CER6, ECERIFERUM 6, CUT1, CUTICULAR 1, G2, KCS6, 3-ketoacyl-CoA synthase 6, POP1, POLLEN-PISTIL INCOMPATIBILITY 1 |
-0.703 |
-0.070 |
| 193 |
263431_at |
AT2G22170
|
PLAT2, PLAT domain protein 2 |
-0.697 |
-0.028 |
| 194 |
257938_at |
AT3G19820
|
CBB1, CABBAGE 1, DIM, DIMINUTIA, DIM1, DIMINUTO 1, DWF1, DWARF 1, EVE1, ENHANCED VERY-LOW-FLUENCE RESPONSES 1 |
-0.683 |
0.001 |
| 195 |
263664_at |
AT1G04250
|
AXR3, AUXIN RESISTANT 3, IAA17, indole-3-acetic acid inducible 17 |
-0.662 |
-0.042 |
| 196 |
254034_at |
AT4G25960
|
ABCB2, ATP-binding cassette B2, PGP2, P-glycoprotein 2 |
-0.658 |
-0.065 |
| 197 |
256825_at |
AT3G22120
|
CWLP, cell wall-plasma membrane linker protein |
-0.651 |
-0.030 |
| 198 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.638 |
0.114 |
| 199 |
252604_at |
AT3G45060
|
ATNRT2.6, ARABIDOPSIS THALIANA HIGH AFFINITY NITRATE TRANSPORTER 2.6, NRT2.6, high affinity nitrate transporter 2.6 |
-0.637 |
-0.146 |
| 200 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.624 |
-0.061 |
| 201 |
259430_at |
AT1G01610
|
ATGPAT4, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 4, GPAT4, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 4 |
-0.623 |
0.013 |
| 202 |
262577_at |
AT1G15290
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.619 |
0.010 |
| 203 |
253372_at |
AT4G33220
|
ATPME44, A. THALIANA PECTIN METHYLESTERASE 44, PME44, pectin methylesterase 44 |
-0.616 |
-0.041 |
| 204 |
253042_at |
AT4G37550
|
[Acetamidase/Formamidase family protein] |
-0.614 |
-0.346 |
| 205 |
246701_at |
AT5G28020
|
ATCYSD2, CYSTEINE SYNTHASE D2, CYSD2, cysteine synthase D2 |
-0.612 |
0.013 |
| 206 |
245845_at |
AT1G26150
|
AtPERK10, proline-rich extensin-like receptor kinase 10, PERK10, proline-rich extensin-like receptor kinase 10 |
-0.595 |
-0.136 |
| 207 |
248419_at |
AT5G51550
|
EXL3, EXORDIUM like 3 |
-0.593 |
-0.070 |
| 208 |
259592_at |
AT1G27950
|
LTPG1, glycosylphosphatidylinositol-anchored lipid protein transfer 1 |
-0.583 |
0.072 |
| 209 |
246313_at |
AT1G31920
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.566 |
-0.030 |
| 210 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.560 |
0.218 |
| 211 |
260140_at |
AT1G66390
|
ATMYB90, MYB90, myb domain protein 90, PAP2, PRODUCTION OF ANTHOCYANIN PIGMENT 2 |
-0.538 |
-0.043 |
| 212 |
248186_at |
AT5G53880
|
unknown |
-0.533 |
0.063 |
| 213 |
254041_at |
AT4G25830
|
[Uncharacterised protein family (UPF0497)] |
-0.527 |
-0.216 |
| 214 |
256379_at |
AT1G66840
|
PMI2, plastid movement impaired 2, WEB2, WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 2 |
-0.518 |
0.047 |
| 215 |
266415_at |
AT2G38530
|
cdf3, cell growth defect factor-3, LP2, LTP2, lipid transfer protein 2 |
-0.517 |
0.117 |
| 216 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
-0.498 |
-0.403 |
| 217 |
257168_at |
AT3G24430
|
HCF101, HIGH-CHLOROPHYLL-FLUORESCENCE 101 |
-0.493 |
-0.123 |
| 218 |
250261_at |
AT5G13400
|
[Major facilitator superfamily protein] |
-0.486 |
-0.071 |
| 219 |
267045_at |
AT2G34180
|
ATWL2, WPL4-LIKE 2, CIPK13, CBL-interacting protein kinase 13, SnRK3.7, SNF1-RELATED PROTEIN KINASE 3.7, WL2, WPL4-LIKE 2 |
-0.484 |
-0.094 |
| 220 |
246339_at |
AT3G44890
|
RPL9, ribosomal protein L9 |
-0.483 |
-0.184 |
| 221 |
265028_at |
AT1G24530
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.476 |
-0.013 |
| 222 |
258468_at |
AT3G06070
|
unknown |
-0.468 |
-0.077 |
| 223 |
259839_at |
AT1G52190
|
AtNPF1.2, NPF1.2, NRT1/ PTR family 1.2, NRT1.11, nitrate transporter 1.11 |
-0.467 |
-0.027 |
| 224 |
260623_at |
AT1G08090
|
ACH1, ATNRT2.1, NITRATE TRANSPORTER 2.1, ATNRT2:1, nitrate transporter 2:1, LIN1, LATERAL ROOT INITIATION 1, NRT2, NITRATE TRANSPORTER 2, NRT2.1, NITRATE TRANSPORTER 2.1, NRT2:1, nitrate transporter 2:1, NRT2;1AT |
-0.464 |
-0.086 |
| 225 |
251317_at |
AT3G61490
|
[Pectin lyase-like superfamily protein] |
-0.459 |
0.053 |
| 226 |
264071_at |
AT2G27920
|
SCPL51, serine carboxypeptidase-like 51 |
-0.454 |
-0.127 |
| 227 |
245877_at |
AT1G26220
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.454 |
-0.096 |
| 228 |
261070_at |
AT1G07390
|
AtRLP1, receptor like protein 1, RLP1, receptor like protein 1 |
-0.448 |
-0.038 |
| 229 |
248634_at |
AT5G49030
|
OVA2, ovule abortion 2 |
-0.448 |
-0.018 |
| 230 |
247563_at |
AT5G61130
|
AtPDCB1, PDCB1, plasmodesmata callose-binding protein 1 |
-0.446 |
-0.020 |
| 231 |
260685_at |
AT1G17650
|
AtGLYR2, GLYR2, glyoxylate reductase 2, GR2, GLYOXYLATE REDUCTASE 2 |
-0.436 |
-0.061 |
| 232 |
257510_at |
AT1G55360
|
unknown |
-0.435 |
0.012 |
| 233 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
-0.431 |
0.021 |
| 234 |
259579_at |
AT1G28010
|
ABCB14, ATP-binding cassette B14, ATABCB14, Arabidopsis thaliana ATP-binding cassette B14, MDR12, multi-drug resistance 12, PGP14, P-glycoprotein 14 |
-0.429 |
-0.025 |
| 235 |
253806_at |
AT4G28270
|
ATRMA2, RMA2, RING membrane-anchor 2 |
-0.428 |
0.004 |
| 236 |
255793_at |
AT2G33250
|
unknown |
-0.424 |
0.056 |
| 237 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
-0.424 |
0.039 |
| 238 |
260890_at |
AT1G29090
|
[Cysteine proteinases superfamily protein] |
-0.423 |
0.175 |
| 239 |
245592_at |
AT4G14540
|
NF-YB3, nuclear factor Y, subunit B3 |
-0.422 |
-0.264 |
| 240 |
255719_at |
AT1G32080
|
AtLrgB, LrgB |
-0.414 |
0.089 |
| 241 |
261375_at |
AT1G53160
|
FTM6, FLORAL TRANSITION AT THE MERISTEM6, SPL4, squamosa promoter binding protein-like 4 |
-0.409 |
-0.011 |
| 242 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
-0.407 |
0.012 |
| 243 |
267472_at |
AT2G02850
|
ARPN, plantacyanin |
-0.406 |
0.004 |
| 244 |
266929_at |
AT2G45850
|
AHL9, AT-hook motif nuclear localized protein 9 |
-0.400 |
-0.136 |
| 245 |
245117_at |
AT2G41560
|
ACA4, autoinhibited Ca(2+)-ATPase, isoform 4 |
-0.398 |
-0.097 |
| 246 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
-0.394 |
-0.028 |
| 247 |
249035_at |
AT5G44190
|
ATGLK2, GLK2, GOLDEN2-like 2, GPRI2, GBF'S PRO-RICH REGION-INTERACTING FACTOR 2 |
-0.389 |
-0.058 |
| 248 |
257516_at |
AT1G69040
|
ACR4, ACT domain repeat 4 |
-0.387 |
0.122 |
| 249 |
246576_at |
AT1G31650
|
ATROPGEF14, ROPGEF14, ROP (rho of plants) guanine nucleotide exchange factor 14 |
-0.386 |
0.061 |
| 250 |
254535_at |
AT4G19710
|
AK-HSDH, ASPARTATE KINASE-HOMOSERINE DEHYDROGENASE, AK-HSDH II, aspartate kinase-homoserine dehydrogenase ii |
-0.383 |
0.084 |
| 251 |
261198_at |
AT1G12940
|
ATNRT2.5, nitrate transporter2.5, NRT2.5, nitrate transporter2.5 |
-0.382 |
0.008 |
| 252 |
263788_at |
AT2G24580
|
[FAD-dependent oxidoreductase family protein] |
-0.381 |
0.020 |
| 253 |
250936_at |
AT5G03120
|
unknown |
-0.381 |
0.213 |
| 254 |
248938_at |
AT5G45780
|
[Leucine-rich repeat protein kinase family protein] |
-0.380 |
-0.009 |
| 255 |
245029_at |
AT2G26580
|
YAB5, YABBY5 |
-0.380 |
0.012 |
| 256 |
256255_at |
AT3G11280
|
[Duplicated homeodomain-like superfamily protein] |
-0.379 |
-0.024 |
| 257 |
253911_at |
AT4G27300
|
[S-locus lectin protein kinase family protein] |
-0.379 |
0.071 |
| 258 |
265993_at |
AT2G24160
|
[pseudogene] |
-0.379 |
0.049 |
| 259 |
260167_at |
AT1G71970
|
unknown |
-0.378 |
-0.015 |
| 260 |
264200_at |
AT1G22650
|
A/N-InvD, alkaline/neutral invertase D |
-0.377 |
-0.040 |
| 261 |
261224_at |
AT1G20160
|
ATSBT5.2 |
-0.377 |
0.284 |
| 262 |
247754_at |
AT5G59080
|
unknown |
-0.376 |
0.027 |
| 263 |
251235_at |
AT3G62860
|
[alpha/beta-Hydrolases superfamily protein] |
-0.374 |
0.005 |
| 264 |
250906_at |
AT5G03650
|
SBE2.2, starch branching enzyme 2.2 |
-0.373 |
-0.128 |
| 265 |
245701_at |
AT5G04140
|
FD-GOGAT, FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE, GLS1, FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE 1, GLU1, glutamate synthase 1, GLUS |
-0.371 |
-0.161 |
| 266 |
252863_at |
AT4G39800
|
ATIPS1, INOSITOL 3-PHOSPHATE SYNTHASE 1, ATMIPS1, MYO-INOSITOL-1-PHOSPHATE SYNTHASE 1, MI-1-P SYNTHASE, MIPS1, D-myo-Inositol 3-Phosphate Synthase 1, MIPS1, myo-inositol-1-phosphate synthase 1 |
-0.359 |
0.012 |
| 267 |
266856_at |
AT2G26910
|
ABCG32, ATP-binding cassette G32, AtABCG32, ATPDR4, PLEIOTROPIC DRUG RESISTANCE 4, PDR4, pleiotropic drug resistance 4, PEC1, PERMEABLE CUTICLE 1 |
-0.355 |
-0.126 |
| 268 |
254798_at |
AT4G13050
|
[Acyl-ACP thioesterase] |
-0.355 |
0.040 |
| 269 |
261046_at |
AT1G01390
|
[UDP-Glycosyltransferase superfamily protein] |
-0.353 |
-0.009 |
| 270 |
267377_at |
AT2G26250
|
FDH, FIDDLEHEAD, KCS10, 3-ketoacyl-CoA synthase 10 |
-0.353 |
0.068 |
| 271 |
264782_at |
AT1G08810
|
AtMYB60, myb domain protein 60, MYB60, myb domain protein 60 |
-0.350 |
0.011 |
| 272 |
247101_at |
AT5G66530
|
[Galactose mutarotase-like superfamily protein] |
-0.348 |
-0.023 |
| 273 |
264466_at |
AT1G10380
|
[Putative membrane lipoprotein] |
-0.347 |
-0.069 |
| 274 |
259116_at |
AT3G01350
|
[Major facilitator superfamily protein] |
-0.347 |
-0.043 |
| 275 |
265823_at |
AT2G35760
|
[Uncharacterised protein family (UPF0497)] |
-0.346 |
-0.084 |
| 276 |
254242_at |
AT4G23200
|
CRK12, cysteine-rich RLK (RECEPTOR-like protein kinase) 12 |
-0.342 |
0.179 |
| 277 |
255104_at |
AT4G08685
|
SAH7 |
-0.334 |
-0.051 |
| 278 |
253218_at |
AT4G34980
|
SLP2, subtilisin-like serine protease 2 |
-0.333 |
-0.020 |
| 279 |
245816_at |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
-0.333 |
-0.015 |
| 280 |
249755_at |
AT5G24580
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.331 |
-0.076 |
| 281 |
256192_at |
AT1G30110
|
ATNUDX25, nudix hydrolase homolog 25, NUDX25, nudix hydrolase homolog 25 |
-0.330 |
0.130 |
| 282 |
259863_at |
AT1G72630
|
ELF4-L2, ELF4-like 2 |
-0.329 |
-0.003 |
| 283 |
264527_at |
AT1G30760
|
[FAD-binding Berberine family protein] |
-0.329 |
0.411 |
| 284 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.328 |
-0.026 |
| 285 |
260170_at |
AT1G71890
|
ATSUC5, SUCROSE-PROTON SYMPORTER 5, SUC5 |
-0.328 |
0.087 |
| 286 |
261598_at |
AT1G49750
|
[Leucine-rich repeat (LRR) family protein] |
-0.327 |
-0.154 |
| 287 |
251154_at |
AT3G63110
|
ATIPT3, isopentenyltransferase 3, IPT3, isopentenyltransferase 3 |
-0.327 |
0.026 |
| 288 |
254901_at |
AT4G11530
|
CRK34, cysteine-rich RLK (RECEPTOR-like protein kinase) 34 |
-0.327 |
0.068 |
| 289 |
250203_at |
AT5G13980
|
[Glycosyl hydrolase family 38 protein] |
-0.322 |
-0.006 |
| 290 |
255913_at |
AT1G66980
|
GDPDL2, Glycerophosphodiester phosphodiesterase (GDPD) like 2, SNC4, suppressor of npr1-1 constitutive 4 |
-0.319 |
0.100 |
| 291 |
246997_at |
AT5G67390
|
unknown |
-0.314 |
0.145 |
| 292 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.314 |
-0.259 |
| 293 |
261881_at |
AT1G80760
|
NIP6, NIP6;1, NOD26-like intrinsic protein 6;1, NLM7 |
-0.314 |
-0.037 |
| 294 |
261189_at |
AT1G33040
|
NACA5, nascent polypeptide-associated complex subunit alpha-like protein 5 |
-0.313 |
0.021 |
| 295 |
252023_at |
AT3G52920
|
unknown |
-0.312 |
-0.064 |
| 296 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
-0.308 |
0.302 |
| 297 |
267414_at |
AT2G34790
|
EDA28, EMBRYO SAC DEVELOPMENT ARREST 28, MEE23, MATERNAL EFFECT EMBRYO ARREST 23 |
-0.308 |
0.093 |
| 298 |
253886_at |
AT4G27710
|
CYP709B3, cytochrome P450, family 709, subfamily B, polypeptide 3 |
-0.306 |
-0.085 |
| 299 |
263410_at |
AT2G04039
|
unknown |
-0.306 |
-0.118 |
| 300 |
260769_at |
AT1G49010
|
[Duplicated homeodomain-like superfamily protein] |
-0.305 |
0.087 |
| 301 |
247643_at |
AT5G60450
|
ARF4, auxin response factor 4 |
-0.303 |
0.037 |
| 302 |
265882_at |
AT2G42490
|
[Copper amine oxidase family protein] |
-0.300 |
0.024 |
| 303 |
265682_at |
AT2G24390
|
[AIG2-like (avirulence induced gene) family protein] |
-0.300 |
0.070 |
| 304 |
257911_at |
AT3G25530
|
ATGHBDH, AtGLYR1, GHBDH, GLYR1, glyoxylate reductase 1, GR1, GLYOXYLATE REDUCTASE 1 |
-0.299 |
0.042 |
| 305 |
247694_at |
AT5G59750
|
AtRIBA3, RIBA3, homolog of ribA 3 |
-0.299 |
-0.027 |
| 306 |
248619_at |
AT5G49630
|
AAP6, amino acid permease 6 |
-0.298 |
0.105 |
| 307 |
262736_at |
AT1G28570
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.297 |
-0.070 |
| 308 |
264313_at |
AT1G70410
|
ATBCA4, BETA CARBONIC ANHYDRASE 4, BCA4, beta carbonic anhydrase 4, CA4, BETA CARBONIC ANHYDRASE 4 |
-0.295 |
-0.010 |
| 309 |
265560_at |
AT2G05520
|
ATGRP-3, GLYCINE-RICH PROTEIN 3, ATGRP3, ARABIDOPSIS GLYCINE RICH PROTEIN 3, GRP-3, glycine-rich protein 3, GRP3, GLYCINE-RICH PROTEIN 3 |
-0.295 |
-0.127 |
| 310 |
255857_at |
AT1G67080
|
ABA4, abscisic acid (ABA)-deficient 4 |
-0.294 |
0.014 |
| 311 |
249932_at |
AT5G22390
|
unknown |
-0.293 |
-0.153 |
| 312 |
262162_at |
AT1G78020
|
unknown |
-0.292 |
-0.104 |
| 313 |
255957_at |
AT1G22160
|
unknown |
-0.291 |
-0.298 |
| 314 |
256222_at |
AT1G56210
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.288 |
-0.045 |
| 315 |
261469_at |
AT1G28340
|
AtRLP4, receptor like protein 4, RLP4, receptor like protein 4 |
-0.287 |
0.018 |
| 316 |
259608_at |
AT1G27960
|
ECT9, evolutionarily conserved C-terminal region 9 |
-0.287 |
-0.089 |
| 317 |
262937_at |
AT1G79560
|
EMB1047, EMBRYO DEFECTIVE 1047, EMB156, EMBRYO DEFECTIVE 156, EMB36, EMBRYO DEFECTIVE 36, FTSH12, FTSH protease 12 |
-0.286 |
0.045 |
| 318 |
263192_at |
AT1G36160
|
ACC1, acetyl-CoA carboxylase 1, AT-ACC1, EMB22, EMBRYO DEFECTIVE 22, GK, GURKE, GSD1, GLOSSYHEAD 1, PAS3, PASTICCINO 3, SFR3, SENSITIVE TO FREEZING 3 |
-0.286 |
0.073 |
| 319 |
258362_at |
AT3G14280
|
unknown |
-0.285 |
0.023 |
| 320 |
246454_at |
AT5G16710
|
DHAR3, dehydroascorbate reductase 1 |
-0.285 |
0.020 |
| 321 |
264613_at |
AT1G04640
|
LIP2, lipoyltransferase 2 |
-0.281 |
0.060 |
| 322 |
262250_at |
AT1G48280
|
[hydroxyproline-rich glycoprotein family protein] |
-0.281 |
-0.184 |
| 323 |
248037_at |
AT5G55930
|
ATOPT1, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 1, OPT1, oligopeptide transporter 1 |
-0.279 |
-0.003 |
| 324 |
266950_at |
AT2G18910
|
[hydroxyproline-rich glycoprotein family protein] |
-0.279 |
0.035 |
| 325 |
249710_at |
AT5G35630
|
ATGSL1, GLUTAMINE SYNTHETASE LIKE 1, GLN2, GLUTAMINE SYNTHETASE 2, GS2, glutamine synthetase 2 |
-0.276 |
0.059 |
| 326 |
266735_at |
AT2G46930
|
[Pectinacetylesterase family protein] |
-0.274 |
-0.010 |
| 327 |
258456_at |
AT3G22420
|
ATWNK2, ARABIDOPSIS THALIANA WITH NO K 2, WNK2, with no lysine (K) kinase 2, ZIK3 |
-0.271 |
-0.038 |
| 328 |
263570_at |
AT2G27150
|
AAO3, abscisic aldehyde oxidase 3, AOdelta, Aldehyde oxidase delta, At-AO3, Arabidopsis thaliana aldehyde oxidase 3, AtAAO3 |
-0.271 |
-0.067 |
| 329 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
-0.269 |
-0.130 |
| 330 |
266038_at |
AT2G07680
|
ABCC13, ATP-binding cassette C13, ATMRP11, multidrug resistance-associated protein 11, MRP11, multidrug resistance-associated protein 11 |
-0.269 |
-0.090 |
| 331 |
264471_at |
AT1G67120
|
AtMDN1, MDN1, MIDASIN 1 |
-0.267 |
0.069 |
| 332 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.266 |
0.046 |
| 333 |
259903_at |
AT1G74160
|
TRM4, TON1 Recruiting Motif 4 |
-0.266 |
-0.001 |
| 334 |
245690_at |
AT5G04230
|
ATPAL3, PAL3, phenyl alanine ammonia-lyase 3 |
-0.265 |
0.100 |
| 335 |
253749_at |
AT4G29080
|
IAA27, indole-3-acetic acid inducible 27, PAP2, phytochrome-associated protein 2 |
-0.265 |
-0.123 |
| 336 |
263909_at |
AT2G36490
|
AtROS1, DML1, demeter-like 1, ROS1, REPRESSOR OF SILENCING1 |
-0.263 |
-0.175 |
| 337 |
256789_at |
AT3G13672
|
SINA2 |
-0.262 |
-0.263 |
| 338 |
252929_at |
AT4G38970
|
AtFBA2, FBA2, fructose-bisphosphate aldolase 2 |
-0.262 |
-0.231 |
| 339 |
264790_at |
AT2G17820
|
AHK1, ATHK1, histidine kinase 1, HK1, histidine kinase 1 |
-0.258 |
0.073 |
| 340 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
-0.258 |
-0.402 |
| 341 |
261307_at |
AT1G48520
|
GATB, GLU-ADT subunit B |
-0.257 |
0.006 |
| 342 |
253225_at |
AT4G35020
|
ARAC3, RAC-like 3, ATROP6, RAC3, RAC-like 3, RHO1PS, ROP6, RHO-RELATED PROTEIN FROM PLANTS 6 |
-0.254 |
-0.053 |
| 343 |
255798_at |
AT2G33255
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.254 |
0.032 |
| 344 |
247628_at |
AT5G60400
|
unknown |
-0.254 |
0.073 |
| 345 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
-0.250 |
0.135 |
| 346 |
267006_at |
AT2G34190
|
[Xanthine/uracil permease family protein] |
-0.249 |
0.019 |
| 347 |
257794_at |
AT3G27050
|
unknown |
-0.249 |
-0.019 |
| 348 |
257547_at |
AT3G13000
|
unknown |
-0.249 |
-0.007 |
| 349 |
262166_at |
AT1G74840
|
[Homeodomain-like superfamily protein] |
-0.247 |
0.017 |
| 350 |
256613_at |
AT3G29290
|
emb2076, embryo defective 2076 |
-0.246 |
-0.045 |
| 351 |
261133_at |
AT1G19715
|
[Mannose-binding lectin superfamily protein] |
-0.245 |
-0.239 |
| 352 |
249059_at |
AT5G44530
|
[Subtilase family protein] |
-0.242 |
0.091 |
| 353 |
265962_at |
AT2G37460
|
UMAMIT12, Usually multiple acids move in and out Transporters 12 |
-0.240 |
-0.134 |
| 354 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.240 |
-0.085 |
| 355 |
262172_at |
AT1G74970
|
RPS9, ribosomal protein S9, TWN3 |
-0.239 |
-0.003 |
| 356 |
252158_at |
AT3G50530
|
CRK, CDPK-related kinase |
-0.239 |
-0.079 |
| 357 |
255310_at |
AT4G04955
|
ALN, allantoinase, ATALN, allantoinase |
-0.238 |
-0.153 |
| 358 |
263947_at |
AT2G35820
|
[ureidoglycolate hydrolases] |
-0.238 |
0.063 |
| 359 |
250610_at |
AT5G07550
|
ATGRP19, GRP19, glycine-rich protein 19 |
-0.237 |
0.031 |
| 360 |
252184_at |
AT3G50660
|
AtDWF4, CLM, CLOMAZONE-RESISTANT, CYP90B1, CYTOCHROME P450 90B1, DWF4, DWARF 4, PSC1, PARTIALLY SUPPRESSING COI1 INSENSITIVITY TO JA 1, SAV1, SHADE AVOIDANCE 1, SNP2, SUPPRESSOR OF NPH4 2 |
-0.237 |
-0.121 |
| 361 |
253043_at |
AT4G37540
|
LBD39, LOB domain-containing protein 39 |
-0.236 |
-0.030 |
| 362 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.233 |
-0.233 |
| 363 |
256542_at |
AT1G42550
|
PMI1, PLASTID MOVEMENT IMPAIRED1 |
-0.233 |
-0.093 |
| 364 |
266782_at |
AT2G29120
|
ATGLR2.7, glutamate receptor 2.7, GLR2.7, GLUTAMATE RECEPTOR 2.7, GLR2.7, glutamate receptor 2.7 |
-0.233 |
0.053 |
| 365 |
247284_at |
AT5G64410
|
ATOPT4, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 4, OPT4, oligopeptide transporter 4 |
-0.233 |
-0.027 |
| 366 |
248890_at |
AT5G46270
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.232 |
-0.096 |
| 367 |
267001_at |
AT2G34470
|
PSKF109, UREG, urease accessory protein G |
-0.232 |
0.048 |
| 368 |
257699_at |
AT3G12780
|
PGK1, phosphoglycerate kinase 1 |
-0.232 |
0.060 |
| 369 |
263809_at |
AT2G04570
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.231 |
0.293 |
| 370 |
250429_at |
AT5G10470
|
KAC1, kinesin like protein for actin based chloroplast movement 1, KCA1, KINESIN CDKA;1 ASSOCIATED 1 |
-0.229 |
-0.096 |
| 371 |
246095_at |
AT5G19310
|
CHR23, chromatin remodeling 23 |
-0.229 |
-0.047 |
| 372 |
248614_at |
AT5G49560
|
[Putative methyltransferase family protein] |
-0.228 |
0.004 |
| 373 |
245993_at |
AT5G20700
|
unknown |
-0.228 |
0.032 |