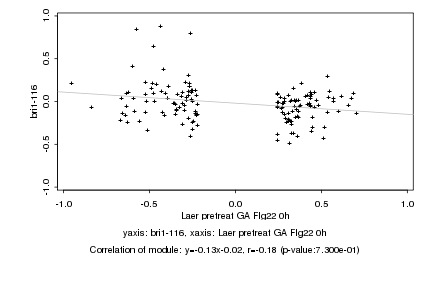
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Laer pretreat GA Flg22 0h |
Log2 signal ratio
bri1-116 |
| 1 |
259036_at |
AT3G09220
|
LAC7, laccase 7 |
0.704 |
-0.129 |
| 2 |
259316_at |
AT3G01175
|
unknown |
0.686 |
0.094 |
| 3 |
260492_at |
AT2G41850
|
ADPG2, ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 2, PGAZAT, polygalacturonase abscission zone A. thaliana |
0.674 |
0.046 |
| 4 |
253060_at |
AT4G37710
|
[VQ motif-containing protein] |
0.654 |
-0.040 |
| 5 |
266232_at |
AT2G02310
|
AtPP2-B6, phloem protein 2-B6, PP2-B6, phloem protein 2-B6 |
0.615 |
0.065 |
| 6 |
265846_at |
AT2G35770
|
scpl28, serine carboxypeptidase-like 28 |
0.597 |
-0.112 |
| 7 |
264425_at |
AT1G61750
|
[Receptor-like protein kinase-related family protein] |
0.569 |
0.001 |
| 8 |
267276_at |
AT2G30130
|
ASL5, LBD12, PCK1, PEACOCK 1 |
0.567 |
0.042 |
| 9 |
265658_at |
AT2G13810
|
ALD1, AGD2-like defense response protein 1, AtALD1, EDTS5, eds two suppressor 5 |
0.543 |
0.122 |
| 10 |
249154_at |
AT5G43410
|
[Integrase-type DNA-binding superfamily protein] |
0.536 |
0.049 |
| 11 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
0.535 |
0.294 |
| 12 |
264543_at |
AT1G55790
|
[FUNCTIONS IN: metal ion binding] |
0.533 |
-0.121 |
| 13 |
246978_at |
AT5G24910
|
CYP714A1, cytochrome P450, family 714, subfamily A, polypeptide 1, ELA1, EUI-like p450 A1 |
0.517 |
-0.300 |
| 14 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
0.507 |
-0.431 |
| 15 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.482 |
-0.042 |
| 16 |
256762_at |
AT3G25655
|
IDL1, inflorescence deficient in abscission (IDA)-like 1 |
0.467 |
0.019 |
| 17 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.458 |
-0.059 |
| 18 |
251281_at |
AT3G61640
|
AGP20, arabinogalactan protein 20, AtAGP20, Arabinogalactan protein 20 |
0.455 |
0.117 |
| 19 |
261814_at |
AT1G08310
|
[alpha/beta-Hydrolases superfamily protein] |
0.444 |
-0.293 |
| 20 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
0.443 |
-0.185 |
| 21 |
261278_at |
AT1G05800
|
DGL, DONGLE |
0.442 |
0.090 |
| 22 |
266271_at |
AT2G29440
|
ATGSTU6, glutathione S-transferase tau 6, GST24, GLUTATHIONE S-TRANSFERASE 24, GSTU6, glutathione S-transferase tau 6 |
0.437 |
-0.339 |
| 23 |
253322_at |
AT4G33980
|
unknown |
0.436 |
-0.046 |
| 24 |
259251_at |
AT3G07600
|
[Heavy metal transport/detoxification superfamily protein ] |
0.434 |
0.114 |
| 25 |
247104_at |
AT5G66620
|
DAR6, DA1-related protein 6 |
0.433 |
-0.051 |
| 26 |
264301_at |
AT1G78780
|
[pathogenesis-related family protein] |
0.432 |
0.066 |
| 27 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
0.432 |
0.043 |
| 28 |
249893_at |
AT5G22555
|
unknown |
0.430 |
-0.016 |
| 29 |
258796_at |
AT3G04630
|
WDL1, WVD2-like 1 |
0.416 |
0.073 |
| 30 |
253259_at |
AT4G34410
|
RRTF1, redox responsive transcription factor 1 |
0.415 |
-0.025 |
| 31 |
255811_at |
AT4G10250
|
ATHSP22.0 |
0.404 |
0.070 |
| 32 |
261700_at |
AT1G32690
|
unknown |
0.382 |
0.211 |
| 33 |
266455_at |
AT2G22760
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.376 |
-0.040 |
| 34 |
259442_at |
AT1G02310
|
MAN1, endo-beta-mannanase 1 |
0.367 |
-0.057 |
| 35 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.366 |
-0.179 |
| 36 |
259780_at |
AT1G29630
|
[5'-3' exonuclease family protein] |
0.365 |
0.022 |
| 37 |
265723_at |
AT2G32140
|
[transmembrane receptors] |
0.364 |
-0.109 |
| 38 |
253100_at |
AT4G37400
|
CYP81F3, cytochrome P450, family 81, subfamily F, polypeptide 3 |
0.358 |
-0.169 |
| 39 |
252537_at |
AT3G45710
|
[Major facilitator superfamily protein] |
0.356 |
-0.398 |
| 40 |
266132_at |
AT2G45130
|
ATSPX3, ARABIDOPSIS THALIANA SPX DOMAIN GENE 3, SPX3, SPX domain gene 3 |
0.355 |
0.020 |
| 41 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
0.350 |
-0.082 |
| 42 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
0.349 |
0.005 |
| 43 |
260179_at |
AT1G70690
|
HWI1, HOPW1-1-INDUCED GENE1, PDLP5, PLASMODESMATA-LOCATED PROTEIN 5 |
0.348 |
-0.183 |
| 44 |
253884_at |
AT4G27670
|
HSP21, heat shock protein 21 |
0.342 |
-0.048 |
| 45 |
260755_at |
AT1G48980
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.338 |
0.017 |
| 46 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
0.335 |
-0.367 |
| 47 |
261109_at |
AT1G75450
|
ATCKX5, ARABIDOPSIS THALIANA CYTOKININ OXIDASE 5, ATCKX6, CYTOKININ OXIDASE 6, CKX5, cytokinin oxidase 5 |
0.335 |
0.157 |
| 48 |
260023_at |
AT1G30040
|
ATGA2OX2, gibberellin 2-oxidase, GA2OX2, gibberellin 2-oxidase, GA2OX2, GIBBERELLIN 2-OXIDASE 2 |
0.331 |
-0.116 |
| 49 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
0.326 |
0.016 |
| 50 |
247208_at |
AT5G64870
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
0.326 |
-0.369 |
| 51 |
265732_at |
AT2G01300
|
unknown |
0.325 |
-0.223 |
| 52 |
252374_at |
AT3G48100
|
ARR5, response regulator 5, ATRR2, ARABIDOPSIS THALIANA RESPONSE REGULATOR 2, IBC6, INDUCED BY CYTOKININ 6, RR5, response regulator 5 |
0.322 |
-0.260 |
| 53 |
264753_at |
AT1G61490
|
[S-locus lectin protein kinase family protein] |
0.317 |
0.005 |
| 54 |
264832_at |
AT1G03660
|
[Ankyrin-repeat containing protein] |
0.315 |
-0.232 |
| 55 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.311 |
-0.212 |
| 56 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
0.310 |
-0.479 |
| 57 |
249836_at |
AT5G23420
|
HMGB6, high-mobility group box 6 |
0.306 |
-0.136 |
| 58 |
255874_at |
AT2G40550
|
ETG1, E2F target gene 1 |
0.306 |
-0.208 |
| 59 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
0.305 |
-0.231 |
| 60 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.304 |
0.074 |
| 61 |
252200_at |
AT3G50280
|
[HXXXD-type acyl-transferase family protein] |
0.295 |
-0.245 |
| 62 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
0.287 |
-0.189 |
| 63 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.284 |
0.038 |
| 64 |
262366_at |
AT1G72890
|
[Disease resistance protein (TIR-NBS class)] |
0.283 |
-0.150 |
| 65 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.280 |
0.008 |
| 66 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.278 |
-0.022 |
| 67 |
248410_at |
AT5G51570
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
0.273 |
-0.064 |
| 68 |
259576_at |
AT1G35330
|
[RING/U-box superfamily protein] |
0.270 |
-0.120 |
| 69 |
253799_at |
AT4G28140
|
[Integrase-type DNA-binding superfamily protein] |
0.268 |
-0.013 |
| 70 |
247691_at |
AT5G59720
|
HSP18.2, heat shock protein 18.2 |
0.265 |
-0.079 |
| 71 |
260917_at |
AT1G02700
|
unknown |
0.260 |
0.058 |
| 72 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
0.245 |
-0.007 |
| 73 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
0.242 |
-0.047 |
| 74 |
264978_at |
AT1G27120
|
[Galactosyltransferase family protein] |
0.241 |
0.097 |
| 75 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
0.241 |
-0.374 |
| 76 |
250024_at |
AT5G18270
|
ANAC087, Arabidopsis NAC domain containing protein 87 |
0.240 |
-0.452 |
| 77 |
247427_at |
AT5G62580
|
[ARM repeat superfamily protein] |
0.239 |
-0.060 |
| 78 |
261838_at |
AT1G16030
|
Hsp70b, heat shock protein 70B |
0.239 |
0.083 |
| 79 |
262061_at |
AT1G80110
|
ATPP2-B11, phloem protein 2-B11, PP2-B11, phloem protein 2-B11 |
0.239 |
-0.011 |
| 80 |
266421_at |
AT2G38540
|
ATLTP1, ARABIDOPSIS THALIANA LIPID TRANSFER PROTEIN 1, LP1, lipid transfer protein 1, LTP1, LIPID TRANSFER PROTEIN 1 |
-0.958 |
0.218 |
| 81 |
263709_at |
AT1G09310
|
unknown |
-0.839 |
-0.069 |
| 82 |
254785_at |
AT4G12730
|
FLA2, FASCICLIN-like arabinogalactan 2 |
-0.673 |
-0.217 |
| 83 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-0.664 |
0.044 |
| 84 |
260267_at |
AT1G68530
|
CER6, ECERIFERUM 6, CUT1, CUTICULAR 1, G2, KCS6, 3-ketoacyl-CoA synthase 6, POP1, POLLEN-PISTIL INCOMPATIBILITY 1 |
-0.662 |
-0.138 |
| 85 |
259592_at |
AT1G27950
|
LTPG1, glycosylphosphatidylinositol-anchored lipid protein transfer 1 |
-0.641 |
-0.161 |
| 86 |
259430_at |
AT1G01610
|
ATGPAT4, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 4, GPAT4, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 4 |
-0.638 |
-0.058 |
| 87 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.636 |
0.099 |
| 88 |
263664_at |
AT1G04250
|
AXR3, AUXIN RESISTANT 3, IAA17, indole-3-acetic acid inducible 17 |
-0.630 |
-0.244 |
| 89 |
260140_at |
AT1G66390
|
ATMYB90, MYB90, myb domain protein 90, PAP2, PRODUCTION OF ANTHOCYANIN PIGMENT 2 |
-0.623 |
0.116 |
| 90 |
262281_at |
AT1G68570
|
AtNPF3.1, NPF3.1, NRT1/ PTR family 3.1 |
-0.601 |
0.420 |
| 91 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.598 |
0.037 |
| 92 |
263431_at |
AT2G22170
|
PLAT2, PLAT domain protein 2 |
-0.593 |
-0.110 |
| 93 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
-0.578 |
0.850 |
| 94 |
253372_at |
AT4G33220
|
ATPME44, A. THALIANA PECTIN METHYLESTERASE 44, PME44, pectin methylesterase 44 |
-0.559 |
-0.225 |
| 95 |
252604_at |
AT3G45060
|
ATNRT2.6, ARABIDOPSIS THALIANA HIGH AFFINITY NITRATE TRANSPORTER 2.6, NRT2.6, high affinity nitrate transporter 2.6 |
-0.527 |
0.091 |
| 96 |
250261_at |
AT5G13400
|
[Major facilitator superfamily protein] |
-0.525 |
-0.127 |
| 97 |
264360_at |
AT1G03310
|
AtBE2, ATISA2, ARABIDOPSIS THALIANA ISOAMYLASE 2, BE2, BRANCHING ENZYME 2, DBE1, debranching enzyme 1, ISA2 |
-0.524 |
0.228 |
| 98 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
-0.519 |
0.006 |
| 99 |
255690_at |
AT4G00360
|
ATT1, ABERRANT INDUCTION OF TYPE THREE 1, CYP86A2, cytochrome P450, family 86, subfamily A, polypeptide 2 |
-0.514 |
-0.334 |
| 100 |
256825_at |
AT3G22120
|
CWLP, cell wall-plasma membrane linker protein |
-0.489 |
0.162 |
| 101 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
-0.485 |
0.220 |
| 102 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
-0.482 |
0.100 |
| 103 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
-0.481 |
0.647 |
| 104 |
266415_at |
AT2G38530
|
cdf3, cell growth defect factor-3, LP2, LTP2, lipid transfer protein 2 |
-0.476 |
0.007 |
| 105 |
251317_at |
AT3G61490
|
[Pectin lyase-like superfamily protein] |
-0.464 |
0.210 |
| 106 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
-0.440 |
0.880 |
| 107 |
267472_at |
AT2G02850
|
ARPN, plantacyanin |
-0.435 |
0.125 |
| 108 |
259579_at |
AT1G28010
|
ABCB14, ATP-binding cassette B14, ATABCB14, Arabidopsis thaliana ATP-binding cassette B14, MDR12, multi-drug resistance 12, PGP14, P-glycoprotein 14 |
-0.425 |
-0.125 |
| 109 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
-0.420 |
0.377 |
| 110 |
254041_at |
AT4G25830
|
[Uncharacterised protein family (UPF0497)] |
-0.415 |
-0.161 |
| 111 |
245816_at |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
-0.407 |
0.096 |
| 112 |
248186_at |
AT5G53880
|
unknown |
-0.400 |
0.041 |
| 113 |
247563_at |
AT5G61130
|
AtPDCB1, PDCB1, plasmodesmata callose-binding protein 1 |
-0.398 |
0.038 |
| 114 |
262563_at |
AT1G34210
|
ATSERK2, SERK2, somatic embryogenesis receptor-like kinase 2 |
-0.393 |
0.176 |
| 115 |
257547_at |
AT3G13000
|
unknown |
-0.364 |
-0.021 |
| 116 |
250936_at |
AT5G03120
|
unknown |
-0.356 |
-0.020 |
| 117 |
266856_at |
AT2G26910
|
ABCG32, ATP-binding cassette G32, AtABCG32, ATPDR4, PLEIOTROPIC DRUG RESISTANCE 4, PDR4, pleiotropic drug resistance 4, PEC1, PERMEABLE CUTICLE 1 |
-0.354 |
-0.033 |
| 118 |
267592_at |
AT2G39710
|
[Eukaryotic aspartyl protease family protein] |
-0.351 |
-0.151 |
| 119 |
253754_at |
AT4G29020
|
[glycine-rich protein] |
-0.348 |
-0.088 |
| 120 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-0.346 |
-0.104 |
| 121 |
251154_at |
AT3G63110
|
ATIPT3, isopentenyltransferase 3, IPT3, isopentenyltransferase 3 |
-0.342 |
0.093 |
| 122 |
254122_at |
AT4G24510
|
CER2, ECERIFERUM 2, VC-2, VC2 |
-0.329 |
-0.060 |
| 123 |
253581_at |
AT4G30660
|
[Low temperature and salt responsive protein family] |
-0.317 |
0.061 |
| 124 |
259608_at |
AT1G27960
|
ECT9, evolutionarily conserved C-terminal region 9 |
-0.311 |
0.110 |
| 125 |
258920_at |
AT3G10520
|
AHB2, haemoglobin 2, ARATH GLB2, ATGLB2, ARABIDOPSIS HEMOGLOBIN 2, GLB2, HEMOGLOBIN 2, HB2, haemoglobin 2, NSHB2, NON-SYMBIOTIC HAEMOGLOBIN 2 |
-0.310 |
-0.019 |
| 126 |
252863_at |
AT4G39800
|
ATIPS1, INOSITOL 3-PHOSPHATE SYNTHASE 1, ATMIPS1, MYO-INOSITOL-1-PHOSPHATE SYNTHASE 1, MI-1-P SYNTHASE, MIPS1, D-myo-Inositol 3-Phosphate Synthase 1, MIPS1, myo-inositol-1-phosphate synthase 1 |
-0.309 |
-0.260 |
| 127 |
257510_at |
AT1G55360
|
unknown |
-0.298 |
-0.100 |
| 128 |
255104_at |
AT4G08685
|
SAH7 |
-0.297 |
-0.035 |
| 129 |
262736_at |
AT1G28570
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.293 |
0.224 |
| 130 |
263192_at |
AT1G36160
|
ACC1, acetyl-CoA carboxylase 1, AT-ACC1, EMB22, EMBRYO DEFECTIVE 22, GK, GURKE, GSD1, GLOSSYHEAD 1, PAS3, PASTICCINO 3, SFR3, SENSITIVE TO FREEZING 3 |
-0.289 |
0.014 |
| 131 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
-0.286 |
0.058 |
| 132 |
258263_at |
AT3G15780
|
unknown |
-0.277 |
0.307 |
| 133 |
262870_at |
AT1G64710
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.276 |
-0.197 |
| 134 |
250610_at |
AT5G07550
|
ATGRP19, GRP19, glycine-rich protein 19 |
-0.276 |
0.079 |
| 135 |
256222_at |
AT1G56210
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.274 |
0.119 |
| 136 |
253749_at |
AT4G29080
|
IAA27, indole-3-acetic acid inducible 27, PAP2, phytochrome-associated protein 2 |
-0.271 |
0.177 |
| 137 |
245029_at |
AT2G26580
|
YAB5, YABBY5 |
-0.271 |
0.185 |
| 138 |
264200_at |
AT1G22650
|
A/N-InvD, alkaline/neutral invertase D |
-0.271 |
0.211 |
| 139 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
-0.266 |
0.799 |
| 140 |
263809_at |
AT2G04570
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.263 |
-0.402 |
| 141 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.261 |
0.109 |
| 142 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.261 |
0.135 |
| 143 |
253911_at |
AT4G27300
|
[S-locus lectin protein kinase family protein] |
-0.260 |
0.029 |
| 144 |
252023_at |
AT3G52920
|
unknown |
-0.253 |
-0.322 |
| 145 |
256255_at |
AT3G11280
|
[Duplicated homeodomain-like superfamily protein] |
-0.253 |
0.010 |
| 146 |
259528_at |
AT1G12330
|
unknown |
-0.251 |
0.136 |
| 147 |
267414_at |
AT2G34790
|
EDA28, EMBRYO SAC DEVELOPMENT ARREST 28, MEE23, MATERNAL EFFECT EMBRYO ARREST 23 |
-0.250 |
-0.242 |
| 148 |
245362_at |
AT4G17460
|
HAT1, JAB, JAIBA |
-0.247 |
-0.226 |
| 149 |
267006_at |
AT2G34190
|
[Xanthine/uracil permease family protein] |
-0.236 |
-0.106 |
| 150 |
254110_at |
AT4G25260
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.235 |
-0.129 |
| 151 |
256192_at |
AT1G30110
|
ATNUDX25, nudix hydrolase homolog 25, NUDX25, nudix hydrolase homolog 25 |
-0.233 |
0.137 |
| 152 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
-0.232 |
0.079 |
| 153 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.229 |
-0.162 |
| 154 |
259903_at |
AT1G74160
|
TRM4, TON1 Recruiting Motif 4 |
-0.228 |
-0.147 |
| 155 |
252033_at |
AT3G51950
|
[Zinc finger (CCCH-type) family protein / RNA recognition motif (RRM)-containing protein] |
-0.226 |
-0.031 |
| 156 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
-0.226 |
-0.273 |
| 157 |
245352_at |
AT4G15490
|
UGT84A3 |
-0.225 |
-0.141 |