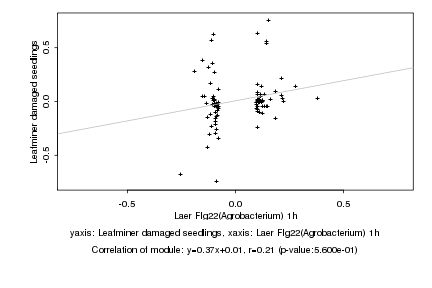
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Laer Flg22(Agrobacterium) 1h |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
263232_at |
AT1G05700
|
[Leucine-rich repeat transmembrane protein kinase protein] |
0.376 |
0.032 |
| 2 |
255111_at |
AT4G08780
|
[Peroxidase superfamily protein] |
0.275 |
0.140 |
| 3 |
260432_at |
AT1G68150
|
ATWRKY9, WRKY9, WRKY DNA-binding protein 9 |
0.218 |
0.008 |
| 4 |
245560_at |
AT4G15480
|
UGT84A1 |
0.214 |
0.032 |
| 5 |
261429_at |
AT1G18860
|
ATWRKY61, WRKY DNA-BINDING PROTEIN 61, WRKY61, WRKY DNA-binding protein 61 |
0.211 |
0.216 |
| 6 |
250987_at |
AT5G02860
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.209 |
0.058 |
| 7 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
0.185 |
-0.157 |
| 8 |
258137_at |
AT3G24515
|
UBC37, ubiquitin-conjugating enzyme 37 |
0.181 |
0.095 |
| 9 |
252698_at |
AT3G43670
|
[Copper amine oxidase family protein] |
0.162 |
0.020 |
| 10 |
262452_at |
AT1G11210
|
unknown |
0.149 |
0.762 |
| 11 |
248605_at |
AT5G49410
|
unknown |
0.148 |
-0.046 |
| 12 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
0.143 |
0.545 |
| 13 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
0.142 |
0.562 |
| 14 |
247531_at |
AT5G61550
|
[U-box domain-containing protein kinase family protein] |
0.140 |
-0.044 |
| 15 |
251489_at |
AT3G59460
|
unknown |
0.134 |
-0.045 |
| 16 |
255860_at |
AT5G34940
|
AtGUS3, glucuronidase 3, GUS3, glucuronidase 3 |
0.133 |
0.074 |
| 17 |
254531_at |
AT4G19650
|
[Mitochondrial transcription termination factor family protein] |
0.125 |
-0.038 |
| 18 |
261599_at |
AT1G49700
|
unknown |
0.123 |
0.002 |
| 19 |
248347_at |
AT5G52250
|
EFO1, EARLY FLOWERING BY OVEREXPRESSION 1, RUP1, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 |
0.121 |
0.015 |
| 20 |
266623_at |
AT2G35390
|
[Phosphoribosyltransferase family protein] |
0.121 |
-0.108 |
| 21 |
264917_at |
AT1G60500
|
DRP4C, Dynamin related protein 4C |
0.121 |
0.018 |
| 22 |
262133_at |
AT1G78000
|
SEL1, SELENATE RESISTANT 1, SULTR1;2, sulfate transporter 1;2 |
0.120 |
-0.007 |
| 23 |
248502_at |
AT5G50450
|
[HCP-like superfamily protein with MYND-type zinc finger] |
0.119 |
0.144 |
| 24 |
245962_at |
AT5G19700
|
[MATE efflux family protein] |
0.114 |
0.002 |
| 25 |
253065_at |
AT4G37740
|
AtGRF2, growth-regulating factor 2, GRF2, growth-regulating factor 2 |
0.113 |
0.049 |
| 26 |
264877_at |
AT2G17330
|
CYP51A1, CYTOCHROME P450 51A1, CYP51G2, CYTOCHROME P450 51G2 |
0.113 |
0.001 |
| 27 |
258177_at |
AT3G21660
|
[UBX domain-containing protein] |
0.112 |
0.072 |
| 28 |
253809_at |
AT4G28320
|
AtMAN5, MAN5, endo-beta-mannase 5 |
0.109 |
-0.008 |
| 29 |
258702_at |
AT3G09730
|
unknown |
0.108 |
-0.093 |
| 30 |
261532_at |
AT1G71680
|
[Transmembrane amino acid transporter family protein] |
0.108 |
-0.094 |
| 31 |
260355_at |
AT1G69180
|
CRC, CRABS CLAW |
0.107 |
0.020 |
| 32 |
259377_at |
AT3G16380
|
PAB6, poly(A) binding protein 6 |
0.105 |
-0.007 |
| 33 |
258434_at |
AT3G16770
|
ATEBP, ethylene-responsive element binding protein, EBP, ethylene-responsive element binding protein, ERF72, ETHYLENE RESPONSE FACTOR 72, RAP2.3, RELATED TO AP2 3 |
0.103 |
0.020 |
| 34 |
249187_at |
AT5G43060
|
RD21B, esponsive to dehydration 21B |
0.102 |
0.092 |
| 35 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
0.101 |
0.636 |
| 36 |
255357_at |
AT4G03930
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.100 |
0.018 |
| 37 |
247641_at |
AT5G60540
|
ATPDX2, PYRIDOXINE BIOSYNTHESIS 2, EMB2407, EMBRYO DEFECTIVE 2407, PDX2, pyridoxine biosynthesis 2 |
0.100 |
0.003 |
| 38 |
246095_at |
AT5G19310
|
CHR23, chromatin remodeling 23 |
0.099 |
-0.238 |
| 39 |
266954_at |
AT2G34530
|
unknown |
0.099 |
-0.088 |
| 40 |
247883_at |
AT5G57790
|
unknown |
0.099 |
0.019 |
| 41 |
265186_at |
AT1G23560
|
unknown |
0.099 |
0.159 |
| 42 |
260458_at |
AT1G68250
|
unknown |
0.098 |
-0.067 |
| 43 |
249151_at |
AT5G43360
|
ATPT4, PHT1;3, phosphate transporter 1;3, PHT3, PHOSPHATE TRANSPORTER 3 |
0.098 |
0.066 |
| 44 |
253541_at |
AT4G31630
|
[Transcriptional factor B3 family protein] |
0.098 |
-0.043 |
| 45 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
0.097 |
-0.005 |
| 46 |
254793_at |
AT4G12930
|
unknown |
0.097 |
-0.019 |
| 47 |
248018_at |
AT5G56470
|
AtGulLO7, GulLO7, L -gulono-1,4-lactone ( L -GulL) oxidase 1 |
0.097 |
-0.001 |
| 48 |
256394_at |
AT3G06290
|
AtSAC3B, SAC3B, yeast Sac3 homolog B |
0.097 |
-0.061 |
| 49 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
-0.257 |
-0.678 |
| 50 |
261088_at |
AT1G07590
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.194 |
0.286 |
| 51 |
252168_at |
AT3G50440
|
ATMES10, ARABIDOPSIS THALIANA METHYL ESTERASE 10, MES10, methyl esterase 10 |
-0.156 |
0.048 |
| 52 |
258516_at |
AT3G06490
|
AtMYB108, myb domain protein 108, BOS1, BOTRYTIS-SUSCEPTIBLE1, MYB108, myb domain protein 108 |
-0.154 |
0.387 |
| 53 |
262284_at |
AT1G68670
|
[myb-like transcription factor family protein] |
-0.144 |
0.048 |
| 54 |
247354_at |
AT5G63590
|
ATFLS3, FLS3, flavonol synthase 3 |
-0.137 |
-0.014 |
| 55 |
262154_at |
AT1G52700
|
[alpha/beta-Hydrolases superfamily protein] |
-0.132 |
-0.146 |
| 56 |
248696_at |
AT5G48360
|
[Actin-binding FH2 (formin homology 2) family protein] |
-0.131 |
-0.420 |
| 57 |
261713_at |
AT1G32640
|
ATMYC2, JAI1, JASMONATE INSENSITIVE 1, JIN1, JASMONATE INSENSITIVE 1, MYC2, RD22BP1, ZBF1 |
-0.125 |
0.323 |
| 58 |
253498_at |
AT4G31890
|
[ARM repeat superfamily protein] |
-0.121 |
-0.303 |
| 59 |
253055_at |
AT4G37630
|
CYCD5;1, cyclin d5;1 |
-0.119 |
0.175 |
| 60 |
258213_at |
AT3G17950
|
unknown |
-0.118 |
-0.120 |
| 61 |
254548_at |
AT4G19865
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.114 |
-0.224 |
| 62 |
260897_at |
AT1G29330
|
AERD2, ARABIDOPSIS ENDOPLASMIC RETICULUM RETENTION DEFECTIVE 2, ATERD2, ARABIDOPSIS THALIANA ENDOPLASMIC RETICULUM RETENTION DEFECTIVE 2, ERD2, ENDOPLASMIC RETICULUM RETENTION DEFECTIVE 2 |
-0.112 |
0.570 |
| 63 |
247973_at |
AT5G56770
|
unknown |
-0.110 |
0.033 |
| 64 |
258189_at |
AT3G17860
|
JAI3, JASMONATE-INSENSITIVE 3, JAZ3, jasmonate-zim-domain protein 3, TIFY6B |
-0.108 |
0.357 |
| 65 |
258317_at |
AT3G22670
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.107 |
-0.019 |
| 66 |
265591_at |
AT2G20150
|
unknown |
-0.106 |
0.054 |
| 67 |
257493_at |
AT1G48180
|
unknown |
-0.105 |
0.010 |
| 68 |
247312_at |
AT5G63970
|
RGLG3, RING DOMAIN LIGASE 3 |
-0.104 |
0.623 |
| 69 |
254369_at |
AT4G21720
|
unknown |
-0.101 |
0.012 |
| 70 |
251600_at |
AT3G57710
|
[Protein kinase superfamily protein] |
-0.100 |
-0.019 |
| 71 |
249053_at |
AT5G44440
|
[FAD-binding Berberine family protein] |
-0.099 |
0.020 |
| 72 |
248660_at |
AT5G48650
|
[Nuclear transport factor 2 (NTF2) family protein with RNA binding (RRM-RBD-RNP motifs) domain] |
-0.098 |
-0.043 |
| 73 |
254017_at |
AT4G26170
|
unknown |
-0.097 |
-0.013 |
| 74 |
251964_at |
AT3G53370
|
[S1FA-like DNA-binding protein] |
-0.097 |
0.274 |
| 75 |
250064_at |
AT5G17890
|
CHS3, CHILLING SENSITIVE 3, DAR4, DA1-related protein 4 |
-0.096 |
-0.181 |
| 76 |
258817_at |
AT3G04750
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.096 |
-0.096 |
| 77 |
261199_at |
AT1G12950
|
RSH2, root hair specific 2 |
-0.095 |
-0.027 |
| 78 |
247056_at |
AT5G66750
|
ATDDM1, CHA1, CHR01, CHROMATIN REMODELING 1, CHR1, chromatin remodeling 1, DDM1, DECREASED DNA METHYLATION 1, SOM1, SOMNIFEROUS 1, SOM4 |
-0.094 |
-0.207 |
| 79 |
245645_at |
AT1G24764
|
ATMAP70-2, microtubule-associated proteins 70-2, MAP70-2, microtubule-associated proteins 70-2 |
-0.094 |
-0.297 |
| 80 |
261775_at |
AT1G76280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.093 |
-0.152 |
| 81 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.092 |
-0.738 |
| 82 |
256703_at |
AT3G30260
|
AGL79, AGAMOUS-like 79 |
-0.090 |
-0.013 |
| 83 |
261373_at |
AT1G53000
|
AtCKS, CKS, CMP-KDO synthetase, KDSB |
-0.090 |
-0.130 |
| 84 |
260010_at |
AT1G68020
|
ATTPS6, TPS6, TREHALOSE -6-PHOSPHATASE SYNTHASE S6 |
-0.090 |
-0.030 |
| 85 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
-0.088 |
-0.257 |
| 86 |
245470_at |
AT4G16020
|
[copia-like retrotransposon family, has a 1.3e-221 P-value blast match to GB:BAA78423 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)GB:BAA78423 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)GB:BAA78423 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)gi|4996361|dbj|BAA78423.1| polyprotein (Arabidopsis thaliana) (Ty1_Copia-element)] |
-0.087 |
-0.025 |
| 87 |
267274_at |
AT2G30160
|
[Mitochondrial substrate carrier family protein] |
-0.087 |
-0.049 |
| 88 |
260677_at |
AT1G07910
|
AtRLG1, AtRLG1, ATRNL, ARABIDOPSIS THALIANA RNA LIGASE, RNL, RNAligase |
-0.087 |
-0.121 |
| 89 |
255246_at |
AT4G05640
|
unknown |
-0.087 |
-0.037 |
| 90 |
250258_at |
AT5G13790
|
AGL15, AGAMOUS-like 15 |
-0.084 |
-0.081 |
| 91 |
259823_at |
AT1G66250
|
[O-Glycosyl hydrolases family 17 protein] |
-0.083 |
-0.342 |
| 92 |
261392_at |
AT1G79780
|
[Uncharacterised protein family (UPF0497)] |
-0.082 |
-0.044 |
| 93 |
265924_at |
AT2G18620
|
[Terpenoid synthases superfamily protein] |
-0.081 |
0.113 |
| 94 |
252245_at |
AT3G49710
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.081 |
-0.060 |
| 95 |
249048_at |
AT5G44300
|
[Dormancy/auxin associated family protein] |
-0.081 |
-0.005 |