|
probeID |
AGICode |
Annotation |
Log2 signal ratio
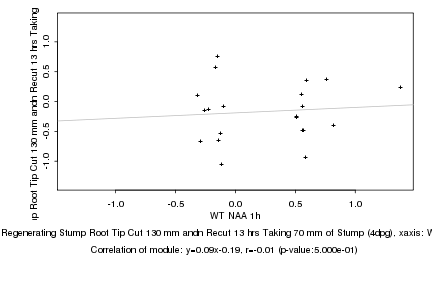
WT NAA 1h |
Log2 signal ratio
Regenerating Stump Root Tip Cut 130 mm andn Recut 13 hrs Taking 70 mm of Stump (4dpg) |
| 1 |
245076_at |
AT2G23170
|
GH3.3 |
1.374 |
0.239 |
| 2 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
0.810 |
-0.400 |
| 3 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
0.759 |
0.377 |
| 4 |
257766_at |
AT3G23030
|
IAA2, indole-3-acetic acid inducible 2 |
0.591 |
0.368 |
| 5 |
248321_at |
AT5G52740
|
[Copper transport protein family] |
0.578 |
-0.932 |
| 6 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
0.561 |
-0.474 |
| 7 |
257900_at |
AT3G28420
|
[Putative membrane lipoprotein] |
0.559 |
-0.083 |
| 8 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
0.557 |
-0.474 |
| 9 |
248801_at |
AT5G47370
|
HAT2 |
0.549 |
0.130 |
| 10 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
0.506 |
-0.254 |
| 11 |
262263_at |
AT1G70940
|
ATPIN3, ARABIDOPSIS PIN-FORMED 3, PIN3, PIN-FORMED 3 |
0.502 |
-0.247 |
| 12 |
261662_at |
AT1G18350
|
ATMKK7, MAP kinase kinase 7, BUD1, BUSHY AND DWARF 1, MKK7, MAP kinase kinase 7, MKK7, MAP KINASE KINASE7 |
-0.322 |
0.106 |
| 13 |
260890_at |
AT1G29090
|
[Cysteine proteinases superfamily protein] |
-0.298 |
-0.659 |
| 14 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.261 |
-0.135 |
| 15 |
260059_at |
AT1G78090
|
ATTPPB, Arabidopsis thaliana trehalose-6-phosphate phosphatase B, TPPB, trehalose-6-phosphate phosphatase B |
-0.230 |
-0.132 |
| 16 |
249211_at |
AT5G42680
|
unknown |
-0.170 |
0.584 |
| 17 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
-0.151 |
0.771 |
| 18 |
248553_at |
AT5G50170
|
[C2 calcium/lipid-binding and GRAM domain containing protein] |
-0.142 |
-0.641 |
| 19 |
260812_at |
AT1G43650
|
UMAMIT22, Usually multiple acids move in and out Transporters 22 |
-0.133 |
-0.523 |
| 20 |
253192_at |
AT4G35370
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.117 |
-1.046 |
| 21 |
262170_at |
AT1G74940
|
unknown |
-0.107 |
-0.077 |