|
probeID |
AGICode |
Annotation |
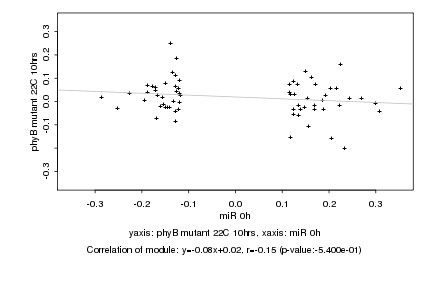
Log2 signal ratio
miR 0h |
Log2 signal ratio
phyB mutant 22C 10hrs |
| 1 |
249614_at |
AT5G37300
|
WSD1 |
0.352 |
0.057 |
| 2 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.306 |
-0.042 |
| 3 |
255654_at |
AT4G00970
|
CRK41, cysteine-rich RLK (RECEPTOR-like protein kinase) 41 |
0.298 |
-0.008 |
| 4 |
265059_at |
AT1G52080
|
AR791 |
0.269 |
0.015 |
| 5 |
247990_at |
AT5G56360
|
PSL4, PRIORITY IN SWEET LIFE 4 |
0.243 |
0.016 |
| 6 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.232 |
-0.199 |
| 7 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
0.224 |
0.163 |
| 8 |
246396_at |
AT1G58180
|
ATBCA6, A. THALIANA BETA CARBONIC ANHYDRASE 6, BCA6, beta carbonic anhydrase 6 |
0.222 |
-0.017 |
| 9 |
266962_at |
AT2G39435
|
TRM18, TON1 Recruiting Motif 18 |
0.214 |
0.056 |
| 10 |
250603_at |
AT5G07820
|
[Plant calmodulin-binding protein-related] |
0.204 |
-0.156 |
| 11 |
262137_at |
AT1G77920
|
TGA7, TGACG sequence-specific binding protein 7 |
0.203 |
0.058 |
| 12 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
0.192 |
0.030 |
| 13 |
245677_at |
AT1G56660
|
unknown |
0.188 |
-0.034 |
| 14 |
264479_at |
AT1G77280
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
0.186 |
0.008 |
| 15 |
260987_at |
AT1G53590
|
NTMC2T6.1, NTMC2TYPE6.1 |
0.171 |
0.077 |
| 16 |
250036_at |
AT5G18340
|
[ARM repeat superfamily protein] |
0.168 |
-0.030 |
| 17 |
249372_at |
AT5G40760
|
G6PD6, glucose-6-phosphate dehydrogenase 6 |
0.168 |
-0.017 |
| 18 |
257073_at |
AT3G19650
|
[cyclin-related] |
0.161 |
0.105 |
| 19 |
250479_at |
AT5G10260
|
AtRABH1e, RAB GTPase homolog H1E, RABH1e, RAB GTPase homolog H1E |
0.155 |
-0.103 |
| 20 |
261538_at |
AT1G01830
|
[ARM repeat superfamily protein] |
0.154 |
0.017 |
| 21 |
258017_at |
AT3G19370
|
unknown |
0.149 |
0.129 |
| 22 |
253227_at |
AT4G35030
|
[Protein kinase superfamily protein] |
0.147 |
-0.022 |
| 23 |
258084_at |
AT3G26020
|
[Protein phosphatase 2A regulatory B subunit family protein] |
0.139 |
-0.033 |
| 24 |
263749_at |
AT2G21520
|
[Sec14p-like phosphatidylinositol transfer family protein] |
0.134 |
-0.056 |
| 25 |
248082_at |
AT5G55400
|
[Actin binding Calponin homology (CH) domain-containing protein] |
0.133 |
-0.014 |
| 26 |
253172_at |
AT4G35060
|
HIPP25, heavy metal associated isoprenylated plant protein 25 |
0.132 |
0.077 |
| 27 |
258291_at |
AT3G23310
|
[AGC (cAMP-dependent, cGMP-dependent and protein kinase C) kinase family protein] |
0.126 |
0.032 |
| 28 |
260363_at |
AT1G70550
|
unknown |
0.124 |
0.086 |
| 29 |
256021_at |
AT1G58270
|
ZW9 |
0.124 |
-0.052 |
| 30 |
261139_at |
AT1G19700
|
BEL10, BEL1-like homeodomain 10, BLH10, BEL1-LIKE HOMEODOMAIN 10 |
0.123 |
-0.032 |
| 31 |
252323_at |
AT3G48530
|
KING1, SNF1-related protein kinase regulatory subunit gamma 1 |
0.117 |
0.031 |
| 32 |
264908_at |
AT2G17440
|
PIRL5, plant intracellular ras group-related LRR 5 |
0.117 |
-0.153 |
| 33 |
264562_at |
AT1G55760
|
[BTB/POZ domain-containing protein] |
0.114 |
0.042 |
| 34 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
0.114 |
0.076 |
| 35 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.288 |
0.018 |
| 36 |
253054_at |
AT4G37580
|
COP3, CONSTITUTIVE PHOTOMORPHOGENIC 3, HLS1, HOOKLESS 1, UNS2, UNUSUAL SUGAR RESPONSE 2 |
-0.254 |
-0.027 |
| 37 |
253480_at |
AT4G31840
|
AtENODL15, ENODL15, early nodulin-like protein 15 |
-0.227 |
0.035 |
| 38 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
-0.195 |
0.007 |
| 39 |
250789_at |
AT5G05630
|
PUT3, POLYAMINE UPTAKE TRANSPORTER 3, RMV1, resistant to methyl viologen 1 |
-0.190 |
0.040 |
| 40 |
257543_at |
AT3G28960
|
[Transmembrane amino acid transporter family protein] |
-0.189 |
0.072 |
| 41 |
263318_at |
AT2G24762
|
AtGDU4, glutamine dumper 4, GDU4, glutamine dumper 4 |
-0.179 |
0.065 |
| 42 |
261893_at |
AT1G80690
|
[PPPDE putative thiol peptidase family protein] |
-0.173 |
0.062 |
| 43 |
259763_at |
AT1G77630
|
LYM3, lysin-motif (LysM) domain protein 3, LYP3, LysM-containing receptor protein 3 |
-0.171 |
0.049 |
| 44 |
252594_at |
AT3G45680
|
[Major facilitator superfamily protein] |
-0.169 |
-0.072 |
| 45 |
247541_at |
AT5G61660
|
[glycine-rich protein] |
-0.168 |
0.030 |
| 46 |
252464_at |
AT3G47160
|
[RING/U-box superfamily protein] |
-0.162 |
-0.018 |
| 47 |
260812_at |
AT1G43650
|
UMAMIT22, Usually multiple acids move in and out Transporters 22 |
-0.156 |
0.021 |
| 48 |
254274_at |
AT4G22770
|
AHL2, AT-hook motif nuclear localized protein 2 |
-0.154 |
-0.012 |
| 49 |
249920_at |
AT5G19260
|
FAF3, FANTASTIC FOUR 3 |
-0.151 |
-0.024 |
| 50 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
-0.150 |
0.080 |
| 51 |
257600_at |
AT3G24770
|
CLE41, CLAVATA3/ESR-RELATED 41 |
-0.147 |
-0.023 |
| 52 |
245383_at |
AT4G17810
|
[C2H2 and C2HC zinc fingers superfamily protein] |
-0.143 |
-0.023 |
| 53 |
261826_at |
AT1G11580
|
ATPMEPCRA, PMEPCRA, methylesterase PCR A |
-0.140 |
0.250 |
| 54 |
262831_at |
AT1G14730
|
[Cytochrome b561/ferric reductase transmembrane protein family] |
-0.135 |
0.126 |
| 55 |
258462_at |
AT3G17350
|
unknown |
-0.134 |
0.001 |
| 56 |
247638_at |
AT5G60490
|
AtFLA12, FLA12, FASCICLIN-like arabinogalactan-protein 12 |
-0.130 |
0.115 |
| 57 |
259837_at |
AT1G52180
|
[Aquaporin-like superfamily protein] |
-0.130 |
-0.039 |
| 58 |
266770_at |
AT2G03090
|
ATEXP15, ATEXPA15, expansin A15, ATHEXP ALPHA 1.3, EXP15, EXPANSIN 15, EXPA15, expansin A15 |
-0.130 |
0.065 |
| 59 |
260298_at |
AT1G80320
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.130 |
-0.083 |
| 60 |
259526_at |
AT1G12570
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-0.126 |
0.187 |
| 61 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
-0.126 |
0.047 |
| 62 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.123 |
0.057 |
| 63 |
265537_at |
AT2G15870
|
[copia-like retrotransposon family, has a 0. P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
-0.122 |
-0.033 |
| 64 |
263305_at |
AT2G01930
|
ATBPC1, BASIC PENTACYSTEINE1, BBR, BPC1, basic pentacysteine1 |
-0.121 |
-0.004 |
| 65 |
245947_at |
AT5G19530
|
ACL5, ACAULIS 5 |
-0.121 |
0.092 |
| 66 |
262543_at |
AT1G34245
|
EPF2, EPIDERMAL PATTERNING FACTOR 2 |
-0.121 |
0.036 |
| 67 |
265689_at |
AT2G24310
|
unknown |
-0.119 |
0.030 |