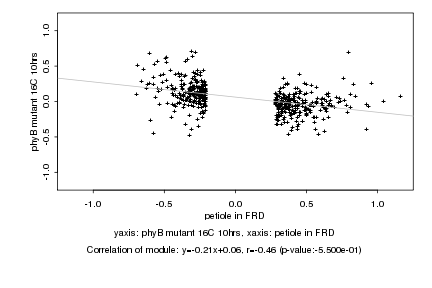
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
petiole in FRD |
Log2 signal ratio
phyB mutant 16C 10hrs |
| 1 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
1.158 |
0.071 |
| 2 |
245076_at |
AT2G23170
|
GH3.3 |
1.039 |
0.011 |
| 3 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
0.952 |
0.256 |
| 4 |
259785_at |
AT1G29490
|
SAUR68, SMALL AUXIN UPREGULATED 68 |
0.929 |
-0.067 |
| 5 |
247474_at |
AT5G62280
|
unknown |
0.915 |
-0.036 |
| 6 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.915 |
-0.381 |
| 7 |
251012_at |
AT5G02580
|
unknown |
0.839 |
0.077 |
| 8 |
250327_at |
AT5G12050
|
unknown |
0.825 |
0.252 |
| 9 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
0.804 |
-0.075 |
| 10 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
0.803 |
0.106 |
| 11 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
0.791 |
0.704 |
| 12 |
259841_at |
AT1G52200
|
[PLAC8 family protein] |
0.782 |
-0.150 |
| 13 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
0.775 |
-0.053 |
| 14 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
0.763 |
0.017 |
| 15 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.755 |
0.337 |
| 16 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
0.735 |
0.003 |
| 17 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
0.726 |
0.050 |
| 18 |
248371_at |
AT5G51810
|
AT2353, ATGA20OX2, GA20OX2, gibberellin 20 oxidase 2 |
0.724 |
-0.012 |
| 19 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
0.704 |
0.063 |
| 20 |
251246_at |
AT3G62100
|
IAA30, indole-3-acetic acid inducible 30 |
0.691 |
-0.077 |
| 21 |
254685_at |
AT4G13790
|
SAUR25, SMALL AUXIN UPREGULATED RNA 25 |
0.673 |
-0.051 |
| 22 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.669 |
-0.065 |
| 23 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.662 |
0.021 |
| 24 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
0.659 |
-0.014 |
| 25 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
0.651 |
0.122 |
| 26 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.651 |
-0.049 |
| 27 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
0.650 |
-0.224 |
| 28 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
0.649 |
0.074 |
| 29 |
259332_at |
AT3G03830
|
SAUR28, SMALL AUXIN UP RNA 28 |
0.638 |
0.011 |
| 30 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
0.637 |
0.102 |
| 31 |
265276_at |
AT2G28400
|
unknown |
0.637 |
-0.094 |
| 32 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
0.634 |
-0.130 |
| 33 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.629 |
-0.088 |
| 34 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
0.627 |
-0.110 |
| 35 |
259443_at |
AT1G02360
|
[Chitinase family protein] |
0.625 |
0.031 |
| 36 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
0.622 |
-0.415 |
| 37 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.614 |
-0.076 |
| 38 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.614 |
-0.242 |
| 39 |
266017_at |
AT2G18690
|
unknown |
0.610 |
-0.020 |
| 40 |
261456_at |
AT1G21050
|
unknown |
0.609 |
0.056 |
| 41 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.607 |
-0.047 |
| 42 |
259479_at |
AT1G19020
|
unknown |
0.602 |
-0.027 |
| 43 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
0.599 |
0.040 |
| 44 |
258100_at |
AT3G23550
|
[MATE efflux family protein] |
0.598 |
-0.116 |
| 45 |
266658_at |
AT2G25735
|
unknown |
0.584 |
-0.457 |
| 46 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
0.577 |
0.049 |
| 47 |
253827_at |
AT4G28085
|
unknown |
0.567 |
0.199 |
| 48 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.566 |
-0.005 |
| 49 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
0.566 |
0.004 |
| 50 |
248123_at |
AT5G54720
|
[Ankyrin repeat family protein] |
0.564 |
-0.017 |
| 51 |
255064_at |
AT4G08950
|
EXO, EXORDIUM |
0.563 |
-0.217 |
| 52 |
252924_at |
AT4G39070
|
BBX20, B-box domain protein 20, BZS1, BZS1 |
0.558 |
-0.384 |
| 53 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
0.554 |
-0.222 |
| 54 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.552 |
-0.294 |
| 55 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
0.545 |
-0.069 |
| 56 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
0.543 |
-0.221 |
| 57 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
0.536 |
-0.009 |
| 58 |
246432_at |
AT5G17490
|
AtRGL3, RGL3, RGA-like protein 3 |
0.534 |
-0.052 |
| 59 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.533 |
0.231 |
| 60 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.528 |
0.130 |
| 61 |
251026_at |
AT5G02200
|
FHL, far-red-elongated hypocotyl1-like |
0.527 |
-0.152 |
| 62 |
256442_at |
AT3G10930
|
unknown |
0.526 |
-0.119 |
| 63 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
0.523 |
-0.053 |
| 64 |
255413_at |
AT4G03140
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.518 |
-0.052 |
| 65 |
251245_at |
AT3G62090
|
PIF6, PHYTOCHROME-INTERACTING FACTOR 6, PIL2, phytochrome interacting factor 3-like 2 |
0.514 |
-0.114 |
| 66 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
0.512 |
-0.110 |
| 67 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
0.498 |
0.123 |
| 68 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
0.497 |
0.253 |
| 69 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.497 |
-0.183 |
| 70 |
245794_at |
AT1G32170
|
XTH30, xyloglucan endotransglucosylase/hydrolase 30, XTR4, xyloglucan endotransglycosylase 4 |
0.496 |
-0.196 |
| 71 |
258541_at |
AT3G07000
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.495 |
-0.086 |
| 72 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
0.495 |
-0.166 |
| 73 |
248801_at |
AT5G47370
|
HAT2 |
0.491 |
-0.074 |
| 74 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
0.491 |
-0.115 |
| 75 |
259729_at |
AT1G77640
|
[Integrase-type DNA-binding superfamily protein] |
0.488 |
-0.004 |
| 76 |
260668_at |
AT1G19530
|
unknown |
0.486 |
0.015 |
| 77 |
257644_at |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
0.486 |
-0.006 |
| 78 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
0.484 |
-0.124 |
| 79 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.483 |
0.262 |
| 80 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
0.480 |
0.104 |
| 81 |
247071_at |
AT5G66640
|
DAR3, DA1-related protein 3 |
0.477 |
-0.026 |
| 82 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
0.470 |
-0.280 |
| 83 |
254410_at |
AT4G21410
|
CRK29, cysteine-rich RLK (RECEPTOR-like protein kinase) 29 |
0.469 |
0.018 |
| 84 |
246108_at |
AT5G28630
|
[glycine-rich protein] |
0.469 |
-0.274 |
| 85 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
0.468 |
-0.066 |
| 86 |
252204_at |
AT3G50340
|
unknown |
0.466 |
-0.015 |
| 87 |
255654_at |
AT4G00970
|
CRK41, cysteine-rich RLK (RECEPTOR-like protein kinase) 41 |
0.456 |
0.154 |
| 88 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.453 |
-0.154 |
| 89 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.450 |
-0.146 |
| 90 |
265732_at |
AT2G01300
|
unknown |
0.450 |
-0.142 |
| 91 |
254063_at |
AT4G25390
|
[Protein kinase superfamily protein] |
0.449 |
0.146 |
| 92 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
0.448 |
-0.190 |
| 93 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
0.447 |
0.382 |
| 94 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.447 |
-0.033 |
| 95 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
0.447 |
0.052 |
| 96 |
262315_at |
AT1G70990
|
[proline-rich family protein] |
0.446 |
0.123 |
| 97 |
247933_at |
AT5G56980
|
unknown |
0.445 |
0.118 |
| 98 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
0.443 |
0.062 |
| 99 |
256190_at |
AT1G30100
|
ATNCED5, NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 5, NCED5, nine-cis-epoxycarotenoid dioxygenase 5 |
0.442 |
-0.033 |
| 100 |
260748_at |
AT1G49210
|
[RING/U-box superfamily protein] |
0.441 |
-0.318 |
| 101 |
250777_at |
AT5G05440
|
PYL5, PYRABACTIN RESISTANCE 1-LIKE 5, RCAR8, regulatory component of ABA receptor 8 |
0.436 |
-0.351 |
| 102 |
262805_at |
AT1G20900
|
AHL27, AT-hook motif nuclear-localized protein 27, ESC, ESCAROLA, ORE7, ORESARA 7 |
0.436 |
-0.007 |
| 103 |
250670_at |
AT5G06860
|
ATPGIP1, POLYGALACTURONASE INHIBITING PROTEIN 1, PGIP1, polygalacturonase inhibiting protein 1 |
0.436 |
-0.224 |
| 104 |
264774_at |
AT1G22890
|
unknown |
0.435 |
-0.089 |
| 105 |
261917_at |
AT1G65920
|
[Regulator of chromosome condensation (RCC1) family with FYVE zinc finger domain] |
0.434 |
0.186 |
| 106 |
261658_at |
AT1G50040
|
unknown |
0.432 |
-0.394 |
| 107 |
248037_at |
AT5G55930
|
ATOPT1, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 1, OPT1, oligopeptide transporter 1 |
0.430 |
-0.058 |
| 108 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
0.427 |
-0.270 |
| 109 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.425 |
0.122 |
| 110 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
0.422 |
0.197 |
| 111 |
265806_at |
AT2G18010
|
SAUR10, SMALL AUXIN UPREGULATED RNA 10 |
0.421 |
-0.049 |
| 112 |
264021_at |
AT2G21200
|
SAUR7, SMALL AUXIN UPREGULATED RNA 7 |
0.421 |
-0.003 |
| 113 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
0.420 |
-0.144 |
| 114 |
253643_at |
AT4G29780
|
unknown |
0.417 |
-0.067 |
| 115 |
266316_at |
AT2G27080
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.417 |
-0.086 |
| 116 |
251752_at |
AT3G55740
|
ATPROT2, PROLINE TRANSPORTER 2, PROT2, proline transporter 2 |
0.417 |
-0.121 |
| 117 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
0.416 |
0.121 |
| 118 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
0.415 |
0.022 |
| 119 |
250108_at |
AT5G15150
|
ATHB-3, homeobox 3, ATHB3, HAT7, HOMEOBOX FROM ARABIDOPSIS THALIANA 7, HB-3, homeobox 3 |
0.414 |
0.077 |
| 120 |
255723_at |
AT3G29575
|
AFP3, ABI five binding protein 3 |
0.412 |
0.017 |
| 121 |
255479_at |
AT4G02380
|
AtLEA5, Arabidopsis thaliana late embryogenensis abundant like 5, SAG21, senescence-associated gene 21 |
0.412 |
-0.168 |
| 122 |
262630_at |
AT1G06520
|
ATGPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, sn-2-GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1 |
0.408 |
0.113 |
| 123 |
264901_at |
AT1G23090
|
AST91, sulfate transporter 91, SULTR3;3 |
0.406 |
-0.129 |
| 124 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.405 |
-0.157 |
| 125 |
249139_at |
AT5G43170
|
AZF3, zinc-finger protein 3, ZF3, zinc-finger protein 3 |
0.404 |
0.051 |
| 126 |
251178_at |
AT3G63440
|
ATCKX6, CYTOKININ OXIDASE 6, ATCKX7, CKX6, cytokinin oxidase/dehydrogenase 6 |
0.402 |
0.017 |
| 127 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
0.401 |
-0.117 |
| 128 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
0.399 |
-0.355 |
| 129 |
245252_at |
AT4G17500
|
ATERF-1, ethylene responsive element binding factor 1, ERF-1, ethylene responsive element binding factor 1 |
0.396 |
-0.060 |
| 130 |
257900_at |
AT3G28420
|
[Putative membrane lipoprotein] |
0.394 |
0.037 |
| 131 |
263207_at |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
0.392 |
-0.404 |
| 132 |
254432_at |
AT4G20830
|
[FAD-binding Berberine family protein] |
0.392 |
-0.234 |
| 133 |
256633_at |
AT3G28340
|
GATL10, galacturonosyltransferase-like 10, GolS8, galactinol synthase 8 |
0.389 |
-0.174 |
| 134 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
0.388 |
-0.156 |
| 135 |
258005_at |
AT3G19390
|
[Granulin repeat cysteine protease family protein] |
0.386 |
0.015 |
| 136 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
0.384 |
-0.012 |
| 137 |
258792_at |
AT3G04640
|
[glycine-rich protein] |
0.381 |
-0.189 |
| 138 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
0.380 |
0.049 |
| 139 |
264551_at |
AT1G09460
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.380 |
-0.105 |
| 140 |
266831_at |
AT2G22830
|
SQE2, squalene epoxidase 2 |
0.376 |
-0.073 |
| 141 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.375 |
-0.047 |
| 142 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.375 |
-0.033 |
| 143 |
254665_at |
AT4G18340
|
[Glycosyl hydrolase superfamily protein] |
0.374 |
-0.143 |
| 144 |
249136_at |
AT5G43180
|
unknown |
0.374 |
0.109 |
| 145 |
260656_at |
AT1G19380
|
unknown |
0.371 |
-0.456 |
| 146 |
247126_at |
AT5G66080
|
APD9, Arabidopsis Pp2c clade D 9 |
0.371 |
0.073 |
| 147 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
0.369 |
-0.023 |
| 148 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
0.369 |
-0.110 |
| 149 |
259566_at |
AT1G20520
|
unknown |
0.368 |
0.037 |
| 150 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
0.368 |
0.008 |
| 151 |
247128_at |
AT5G66110
|
HIPP27, heavy metal associated isoprenylated plant protein 27 |
0.367 |
-0.071 |
| 152 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.367 |
-0.019 |
| 153 |
252862_at |
AT4G39830
|
[Cupredoxin superfamily protein] |
0.365 |
-0.285 |
| 154 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
0.365 |
0.012 |
| 155 |
265877_at |
AT2G42380
|
ATBZIP34, BZIP34 |
0.364 |
0.243 |
| 156 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
0.362 |
0.258 |
| 157 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
0.359 |
0.061 |
| 158 |
248539_at |
AT5G50130
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.358 |
-0.059 |
| 159 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
0.357 |
-0.091 |
| 160 |
247177_at |
AT5G65300
|
unknown |
0.353 |
-0.001 |
| 161 |
252993_at |
AT4G38540
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.352 |
0.030 |
| 162 |
266611_at |
AT2G14960
|
GH3.1 |
0.350 |
-0.031 |
| 163 |
257650_at |
AT3G16800
|
[Protein phosphatase 2C family protein] |
0.349 |
0.047 |
| 164 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
0.347 |
-0.062 |
| 165 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
0.347 |
0.253 |
| 166 |
263565_at |
AT2G15390
|
atfut4, FUT4, fucosyltransferase 4 |
0.347 |
-0.146 |
| 167 |
266814_at |
AT2G44910
|
ATHB-4, ARABIDOPSIS THALIANA HOMEOBOX-LEUCINE ZIPPER PROTEIN 4, ATHB4, homeobox-leucine zipper protein 4, HB4, homeobox-leucine zipper protein 4 |
0.347 |
-0.088 |
| 168 |
267246_at |
AT2G30250
|
ATWRKY25, WRKY25, WRKY DNA-binding protein 25 |
0.342 |
-0.214 |
| 169 |
251507_at |
AT3G59080
|
[Eukaryotic aspartyl protease family protein] |
0.341 |
-0.288 |
| 170 |
260411_at |
AT1G69890
|
unknown |
0.340 |
-0.135 |
| 171 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
0.338 |
0.086 |
| 172 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
0.338 |
-0.041 |
| 173 |
258253_at |
AT3G26760
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.338 |
-0.080 |
| 174 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
0.337 |
0.007 |
| 175 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.337 |
0.113 |
| 176 |
265648_at |
AT2G27500
|
[Glycosyl hydrolase superfamily protein] |
0.336 |
-0.243 |
| 177 |
251870_at |
AT3G54510
|
[Early-responsive to dehydration stress protein (ERD4)] |
0.335 |
-0.097 |
| 178 |
264580_at |
AT1G05340
|
unknown |
0.333 |
0.329 |
| 179 |
245784_at |
AT1G32190
|
[alpha/beta-Hydrolases superfamily protein] |
0.333 |
0.061 |
| 180 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
0.332 |
0.204 |
| 181 |
265547_at |
AT2G28305
|
ATLOG1, LOG1, LONELY GUY 1 |
0.327 |
0.008 |
| 182 |
250267_at |
AT5G12930
|
unknown |
0.327 |
-0.096 |
| 183 |
262114_at |
AT1G02860
|
BAH1, BENZOIC ACID HYPERSENSITIVE 1, NLA, nitrogen limitation adaptation |
0.326 |
-0.127 |
| 184 |
261768_at |
AT1G15550
|
ATGA3OX1, ARABIDOPSIS THALIANA GIBBERELLIN 3 BETA-HYDROXYLASE 1, GA3OX1, gibberellin 3-oxidase 1, GA4, GA REQUIRING 4 |
0.326 |
-0.003 |
| 185 |
247280_at |
AT5G64260
|
EXL2, EXORDIUM like 2 |
0.323 |
-0.137 |
| 186 |
265144_at |
AT1G51170
|
AGC2-3, AGC2 kinase 3, UCN, UNICORN |
0.323 |
0.006 |
| 187 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
0.322 |
-0.321 |
| 188 |
262525_at |
AT1G17060
|
CHI2, CHIBI 2, CYP72C1, cytochrome p450 72c1, SHK1, SHRINK 1, SOB7, SUPPRESSOR OF PHYB-4 7 |
0.320 |
-0.185 |
| 189 |
251763_at |
AT3G55730
|
AtMYB109, myb domain protein 109, MYB109, myb domain protein 109 |
0.320 |
0.045 |
| 190 |
261027_at |
AT1G01340
|
ACBK1, ATCNGC10, cyclic nucleotide gated channel 10, CNGC10, cyclic nucleotide gated channel 10 |
0.320 |
-0.111 |
| 191 |
260819_at |
AT1G06850
|
AtbZIP52, basic leucine-zipper 52, bZIP52, basic leucine-zipper 52 |
0.320 |
-0.036 |
| 192 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
0.320 |
-0.160 |
| 193 |
264836_at |
AT1G03610
|
unknown |
0.317 |
0.027 |
| 194 |
258764_at |
AT3G10720
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.317 |
-0.040 |
| 195 |
256787_at |
AT3G13790
|
ATBFRUCT1, ATCWINV1, ARABIDOPSIS THALIANA CELL WALL INVERTASE 1 |
0.316 |
-0.050 |
| 196 |
251423_at |
AT3G60550
|
CYCP3;2, cyclin p3;2 |
0.315 |
-0.043 |
| 197 |
250657_at |
AT5G07000
|
ATST2B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2B, ST2B, sulfotransferase 2B |
0.314 |
0.188 |
| 198 |
252183_at |
AT3G50740
|
UGT72E1, UDP-glucosyl transferase 72E1 |
0.313 |
0.118 |
| 199 |
249361_at |
AT5G40540
|
[Protein kinase superfamily protein] |
0.312 |
-0.022 |
| 200 |
247743_at |
AT5G59010
|
BSK5, brassinosteroid-signaling kinase 5 |
0.310 |
-0.063 |
| 201 |
246018_at |
AT5G10695
|
unknown |
0.308 |
-0.205 |
| 202 |
258540_at |
AT3G06990
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.306 |
0.143 |
| 203 |
258075_at |
AT3G25900
|
ATHMT-1, HMT-1 |
0.304 |
-0.033 |
| 204 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
0.304 |
0.082 |
| 205 |
266346_at |
AT2G01430
|
ATHB-17, ARABIDOPSIS THALIANA HOMEOBOX-LEUCINE ZIPPER PROTEIN 17, ATHB17, homeobox-leucine zipper protein 17, HB17, homeobox-leucine zipper protein 17 |
0.301 |
-0.034 |
| 206 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
0.301 |
-0.144 |
| 207 |
258953_at |
AT3G01430
|
unknown |
0.301 |
-0.172 |
| 208 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.301 |
-0.074 |
| 209 |
256799_at |
AT3G18560
|
unknown |
0.298 |
-0.109 |
| 210 |
254409_at |
AT4G21400
|
CRK28, cysteine-rich RLK (RECEPTOR-like protein kinase) 28 |
0.297 |
-0.089 |
| 211 |
246943_at |
AT5G25440
|
[Protein kinase superfamily protein] |
0.297 |
-0.266 |
| 212 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
0.296 |
-0.065 |
| 213 |
257766_at |
AT3G23030
|
IAA2, indole-3-acetic acid inducible 2 |
0.295 |
0.096 |
| 214 |
267393_at |
AT2G44500
|
[O-fucosyltransferase family protein] |
0.295 |
-0.320 |
| 215 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
0.293 |
-0.214 |
| 216 |
252879_at |
AT4G39390
|
ATNST-KT1, A. THALIANA NUCLEOTIDE SUGAR TRANSPORTER-KT 1, NST-K1, nucleotide sugar transporter-KT 1 |
0.292 |
-0.058 |
| 217 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
0.292 |
-0.268 |
| 218 |
259596_at |
AT1G28130
|
GH3.17 |
0.290 |
0.032 |
| 219 |
245084_at |
AT2G23290
|
AtMYB70, myb domain protein 70, MYB70, myb domain protein 70 |
0.289 |
0.007 |
| 220 |
250323_at |
AT5G12880
|
[proline-rich family protein] |
0.286 |
0.103 |
| 221 |
266606_at |
AT2G46310
|
CRF5, cytokinin response factor 5 |
0.285 |
0.058 |
| 222 |
261451_at |
AT1G21060
|
unknown |
0.285 |
-0.030 |
| 223 |
255742_at |
AT1G25560
|
AtTEM1, EDF1, ETHYLENE RESPONSE DNA BINDING FACTOR 1, TEM1, TEMPRANILLO 1 |
0.285 |
-0.242 |
| 224 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
0.285 |
-0.313 |
| 225 |
251925_at |
AT3G54000
|
unknown |
0.284 |
0.002 |
| 226 |
262756_at |
AT1G16370
|
ATOCT6, ARABIDOPSIS THALIANA ORGANIC CATION/CARNITINE TRANSPORTER 6, OCT6, organic cation/carnitine transporter 6 |
0.283 |
0.018 |
| 227 |
264512_at |
AT1G09575
|
unknown |
0.283 |
-0.191 |
| 228 |
253791_at |
AT4G28640
|
IAA11, indole-3-acetic acid inducible 11 |
0.280 |
0.026 |
| 229 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.280 |
-0.002 |
| 230 |
264663_at |
AT1G09970
|
LRR XI-23, RLK7, receptor-like kinase 7 |
0.279 |
-0.042 |
| 231 |
257206_at |
AT3G16530
|
[Legume lectin family protein] |
0.279 |
0.121 |
| 232 |
251643_at |
AT3G57550
|
AGK2, guanylate kinase, GK-2, GUANYLATE KINAS 2 |
0.278 |
-0.027 |
| 233 |
254408_at |
AT4G21390
|
B120 |
0.277 |
0.037 |
| 234 |
254371_at |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
-0.702 |
0.102 |
| 235 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.695 |
0.510 |
| 236 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.664 |
0.285 |
| 237 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
-0.647 |
0.455 |
| 238 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
-0.631 |
0.184 |
| 239 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.620 |
0.249 |
| 240 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
-0.610 |
0.258 |
| 241 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-0.607 |
0.680 |
| 242 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.603 |
-0.262 |
| 243 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
-0.583 |
-0.443 |
| 244 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
-0.581 |
0.250 |
| 245 |
252010_at |
AT3G52740
|
unknown |
-0.577 |
0.353 |
| 246 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
-0.570 |
0.531 |
| 247 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.566 |
0.069 |
| 248 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.555 |
0.198 |
| 249 |
265892_at |
AT2G15020
|
unknown |
-0.550 |
0.579 |
| 250 |
248961_at |
AT5G45650
|
[subtilase family protein] |
-0.547 |
0.151 |
| 251 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.539 |
-0.038 |
| 252 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
-0.529 |
0.378 |
| 253 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.523 |
0.181 |
| 254 |
255016_at |
AT4G10120
|
ATSPS4F |
-0.514 |
0.276 |
| 255 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.506 |
0.385 |
| 256 |
256096_at |
AT1G13650
|
unknown |
-0.492 |
0.611 |
| 257 |
252661_at |
AT3G44450
|
unknown |
-0.486 |
0.564 |
| 258 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.486 |
0.635 |
| 259 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
-0.486 |
0.304 |
| 260 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
-0.481 |
-0.014 |
| 261 |
266572_at |
AT2G23840
|
[HNH endonuclease] |
-0.479 |
0.200 |
| 262 |
251058_at |
AT5G01790
|
unknown |
-0.463 |
0.440 |
| 263 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.454 |
0.235 |
| 264 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-0.450 |
-0.213 |
| 265 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.449 |
0.116 |
| 266 |
246142_at |
AT5G19970
|
unknown |
-0.448 |
-0.033 |
| 267 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.445 |
0.179 |
| 268 |
261819_at |
AT1G11410
|
[S-locus lectin protein kinase family protein] |
-0.444 |
0.188 |
| 269 |
247486_at |
AT5G62140
|
unknown |
-0.442 |
0.087 |
| 270 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.436 |
0.172 |
| 271 |
261221_at |
AT1G19960
|
unknown |
-0.431 |
-0.132 |
| 272 |
267505_at |
AT2G45560
|
CYP76C1, cytochrome P450, family 76, subfamily C, polypeptide 1 |
-0.431 |
0.160 |
| 273 |
252495_at |
AT3G46770
|
[AP2/B3-like transcriptional factor family protein] |
-0.430 |
-0.047 |
| 274 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.427 |
-0.059 |
| 275 |
254186_at |
AT4G24010
|
ATCSLG1, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G1, CSLG1, cellulose synthase like G1 |
-0.426 |
0.087 |
| 276 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.426 |
0.385 |
| 277 |
256386_at |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
-0.424 |
-0.059 |
| 278 |
247323_at |
AT5G64170
|
LNK1, night light-inducible and clock-regulated 1 |
-0.423 |
0.193 |
| 279 |
249047_at |
AT5G44410
|
[FAD-binding Berberine family protein] |
-0.409 |
-0.018 |
| 280 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.408 |
0.058 |
| 281 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
-0.407 |
0.398 |
| 282 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
-0.400 |
0.357 |
| 283 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
-0.399 |
-0.023 |
| 284 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
-0.399 |
0.125 |
| 285 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
-0.397 |
-0.017 |
| 286 |
258982_at |
AT3G08870
|
LecRK-VI.1, L-type lectin receptor kinase VI.1 |
-0.397 |
-0.008 |
| 287 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
-0.392 |
0.049 |
| 288 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
-0.391 |
0.087 |
| 289 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.383 |
0.253 |
| 290 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
-0.381 |
0.393 |
| 291 |
245749_at |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.381 |
0.299 |
| 292 |
247467_at |
AT5G62130
|
[Per1-like family protein] |
-0.380 |
0.213 |
| 293 |
247602_at |
AT5G60900
|
RLK1, receptor-like protein kinase 1 |
-0.380 |
0.040 |
| 294 |
257129_at |
AT3G20100
|
CYP705A19, cytochrome P450, family 705, subfamily A, polypeptide 19 |
-0.376 |
0.188 |
| 295 |
246947_at |
AT5G25120
|
CYP71B11, ytochrome p450, family 71, subfamily B, polypeptide 11 |
-0.375 |
0.183 |
| 296 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
-0.371 |
0.077 |
| 297 |
258562_at |
AT3G05980
|
unknown |
-0.370 |
-0.072 |
| 298 |
264271_at |
AT1G60270
|
BGLU6, beta glucosidase 6 |
-0.369 |
0.243 |
| 299 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
-0.369 |
0.091 |
| 300 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
-0.367 |
0.022 |
| 301 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.366 |
0.112 |
| 302 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
-0.366 |
0.082 |
| 303 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.362 |
0.366 |
| 304 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
-0.359 |
0.248 |
| 305 |
262196_at |
AT1G77870
|
MUB5, membrane-anchored ubiquitin-fold protein 5 precursor |
-0.359 |
-0.034 |
| 306 |
254145_at |
AT4G24700
|
unknown |
-0.357 |
0.370 |
| 307 |
261247_at |
AT1G20070
|
unknown |
-0.355 |
0.579 |
| 308 |
260081_at |
AT1G78170
|
unknown |
-0.353 |
0.022 |
| 309 |
257272_at |
AT3G28130
|
UMAMIT44, Usually multiple acids move in and out Transporters 44 |
-0.353 |
0.135 |
| 310 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
-0.352 |
-0.311 |
| 311 |
252411_at |
AT3G47430
|
PEX11B, peroxin 11B |
-0.347 |
0.024 |
| 312 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
-0.344 |
0.598 |
| 313 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
-0.343 |
0.051 |
| 314 |
256509_at |
AT1G75300
|
[NmrA-like negative transcriptional regulator family protein] |
-0.341 |
0.058 |
| 315 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
-0.336 |
0.123 |
| 316 |
245304_at |
AT4G15630
|
[Uncharacterised protein family (UPF0497)] |
-0.335 |
-0.044 |
| 317 |
254662_at |
AT4G18270
|
ATTRANS11, ARABIDOPSIS THALIANA TRANSLOCASE 11, TRANS11, translocase 11 |
-0.332 |
0.143 |
| 318 |
256293_at |
AT1G69440
|
AGO7, ARGONAUTE7, ZIP, ZIPPY |
-0.332 |
0.119 |
| 319 |
261407_at |
AT1G18810
|
[phytochrome kinase substrate-related] |
-0.330 |
0.172 |
| 320 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.330 |
-0.468 |
| 321 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
-0.328 |
0.146 |
| 322 |
250985_at |
AT5G02830
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.327 |
0.193 |
| 323 |
248075_at |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
-0.327 |
0.077 |
| 324 |
256427_at |
AT3G11090
|
LBD21, LOB domain-containing protein 21 |
-0.325 |
0.014 |
| 325 |
254416_at |
AT4G21380
|
ARK3, receptor kinase 3, RK3, receptor kinase 3 |
-0.325 |
-0.190 |
| 326 |
251342_at |
AT3G60690
|
SAUR59, SMALL AUXIN UPREGULATED RNA 59 |
-0.323 |
0.001 |
| 327 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
-0.322 |
0.204 |
| 328 |
247827_at |
AT5G58500
|
LSH5, LIGHT SENSITIVE HYPOCOTYLS 5 |
-0.319 |
0.040 |
| 329 |
247946_at |
AT5G57180
|
CIA2, chloroplast import apparatus 2 |
-0.318 |
0.110 |
| 330 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.317 |
0.260 |
| 331 |
246986_at |
AT5G67280
|
RLK, receptor-like kinase |
-0.317 |
0.110 |
| 332 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
-0.315 |
0.709 |
| 333 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
-0.315 |
-0.389 |
| 334 |
256379_at |
AT1G66840
|
PMI2, plastid movement impaired 2, WEB2, WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 2 |
-0.314 |
0.167 |
| 335 |
261921_at |
AT1G65900
|
unknown |
-0.313 |
-0.046 |
| 336 |
265842_at |
AT2G35700
|
ATERF38, ERF FAMILY PROTEIN 38, ERF38, ERF family protein 38 |
-0.312 |
0.157 |
| 337 |
260773_at |
AT1G78440
|
ATGA2OX1, Arabidopsis thaliana gibberellin 2-oxidase 1, GA2OX1, gibberellin 2-oxidase 1 |
-0.311 |
0.273 |
| 338 |
246235_at |
AT4G36830
|
HOS3-1 |
-0.309 |
0.210 |
| 339 |
251060_at |
AT5G01820
|
ATCIPK14, ATSR1, serine/threonine protein kinase 1, CIPK14, CBL-INTERACTING PROTEIN KINASE 14, PKS24, SOS2-like protein kinase 24, SnRK3.15, SNF1-RELATED PROTEIN KINASE 3.15, SR1, serine/threonine protein kinase 1 |
-0.308 |
0.645 |
| 340 |
258181_at |
AT3G21670
|
AtNPF6.4, NPF6.4, NRT1/ PTR family 6.4 |
-0.305 |
0.330 |
| 341 |
264583_at |
AT1G05170
|
[Galactosyltransferase family protein] |
-0.305 |
0.222 |
| 342 |
250420_at |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
-0.305 |
0.127 |
| 343 |
247977_at |
AT5G56850
|
unknown |
-0.301 |
0.111 |
| 344 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.301 |
0.415 |
| 345 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
-0.299 |
0.213 |
| 346 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
-0.297 |
0.180 |
| 347 |
262411_at |
AT1G34640
|
[peptidases] |
-0.297 |
-0.068 |
| 348 |
259308_at |
AT3G05180
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.296 |
0.086 |
| 349 |
260262_at |
AT1G68470
|
[Exostosin family protein] |
-0.296 |
-0.009 |
| 350 |
252911_at |
AT4G39510
|
CYP96A12, cytochrome P450, family 96, subfamily A, polypeptide 12 |
-0.293 |
0.194 |
| 351 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
-0.293 |
0.001 |
| 352 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.293 |
0.017 |
| 353 |
258033_at |
AT3G21250
|
ABCC8, ATP-binding cassette C8, ATMRP6, ARABIDOPSIS THALIANA MULTIDRUG RESISTANCE-ASSOCIATED PROTEIN 6, MRP6, multidrug resistance-associated protein 6 |
-0.291 |
0.219 |
| 354 |
260603_at |
AT1G55960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.291 |
0.361 |
| 355 |
250859_at |
AT5G04660
|
CYP77A4, cytochrome P450, family 77, subfamily A, polypeptide 4 |
-0.289 |
0.123 |
| 356 |
263128_at |
AT1G78600
|
BBX22, B-box domain protein 22, DBB3, DOUBLE B-BOX 3, LZF1, light-regulated zinc finger protein 1, STH3, SALT TOLERANCE HOMOLOG 3 |
-0.289 |
0.129 |
| 357 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
-0.289 |
0.073 |
| 358 |
246885_at |
AT5G26230
|
MAKR1, membrane-associated kinase regulator 1 |
-0.286 |
0.059 |
| 359 |
260717_at |
AT1G48120
|
[hydrolases] |
-0.284 |
0.319 |
| 360 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
-0.283 |
0.402 |
| 361 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
-0.282 |
0.695 |
| 362 |
263796_at |
AT2G24540
|
AFR, ATTENUATED FAR-RED RESPONSE |
-0.281 |
0.306 |
| 363 |
251826_at |
AT3G55110
|
ABCG18, ATP-binding cassette G18 |
-0.279 |
0.271 |
| 364 |
257748_at |
AT3G18710
|
ATPUB29, ARABIDOPSIS THALIANA PLANT U-BOX 29, PUB29, plant U-box 29 |
-0.279 |
0.225 |
| 365 |
259432_at |
AT1G01520
|
ASG4, ALTERED SEED GERMINATION 4 |
-0.276 |
0.106 |
| 366 |
255032_at |
AT4G09500
|
[UDP-Glycosyltransferase superfamily protein] |
-0.276 |
0.052 |
| 367 |
255294_at |
AT4G04750
|
[Major facilitator superfamily protein] |
-0.275 |
0.110 |
| 368 |
263599_at |
AT2G01830
|
AHK4, ARABIDOPSIS HISTIDINE KINASE 4, ATCRE1, CRE1, CYTOKININ RESPONSE 1, WOL, WOODEN LEG, WOL1, WOODEN LEG 1 |
-0.274 |
0.112 |
| 369 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
-0.274 |
-0.021 |
| 370 |
262526_at |
AT1G17050
|
AtSPS2, SPS2, solanesyl diphosphate synthase 2 |
-0.272 |
0.273 |
| 371 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.272 |
0.148 |
| 372 |
262882_at |
AT1G64900
|
CYP89, CYP89A2, cytochrome P450, family 89, subfamily A, polypeptide 2 |
-0.271 |
0.392 |
| 373 |
253254_at |
AT4G34650
|
SQS2, squalene synthase 2 |
-0.271 |
0.206 |
| 374 |
261318_at |
AT1G53035
|
unknown |
-0.271 |
-0.243 |
| 375 |
263473_at |
AT2G31750
|
UGT74D1, UDP-glucosyl transferase 74D1 |
-0.270 |
0.292 |
| 376 |
264057_at |
AT2G28550
|
RAP2.7, related to AP2.7, TOE1, TARGET OF EARLY ACTIVATION TAGGED (EAT) 1 |
-0.267 |
0.153 |
| 377 |
264102_at |
AT1G79270
|
ECT8, evolutionarily conserved C-terminal region 8 |
-0.267 |
0.442 |
| 378 |
260618_at |
AT1G53230
|
TCP3, TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 3 |
-0.265 |
-0.079 |
| 379 |
265828_at |
AT2G14520
|
unknown |
-0.265 |
0.010 |
| 380 |
267639_at |
AT2G42200
|
AtSPL9, SPL9, squamosa promoter binding protein-like 9 |
-0.264 |
0.123 |
| 381 |
255622_at |
AT4G01070
|
GT72B1, UGT72B1, UDP-GLUCOSE-DEPENDENT GLUCOSYLTRANSFERASE 72 B1 |
-0.263 |
0.098 |
| 382 |
247179_at |
AT5G65320
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.263 |
-0.024 |
| 383 |
255913_at |
AT1G66980
|
GDPDL2, Glycerophosphodiester phosphodiesterase (GDPD) like 2, SNC4, suppressor of npr1-1 constitutive 4 |
-0.263 |
-0.341 |
| 384 |
259042_at |
AT3G03450
|
RGL2, RGA-like 2 |
-0.260 |
0.211 |
| 385 |
266384_at |
AT2G14660
|
unknown |
-0.259 |
0.013 |
| 386 |
246468_at |
AT5G17050
|
UGT78D2, UDP-glucosyl transferase 78D2 |
-0.259 |
0.140 |
| 387 |
250372_at |
AT5G11460
|
unknown |
-0.255 |
0.144 |
| 388 |
251827_at |
AT3G55120
|
A11, AtCHI, CFI, CHALCONE FLAVANONE ISOMERASE, CHI, chalcone isomerase, TT5, TRANSPARENT TESTA 5 |
-0.255 |
0.080 |
| 389 |
248625_at |
AT5G48880
|
KAT5, 3-KETO-ACYL-COENZYME A THIOLASE 5, PKT1, PEROXISOMAL-3-KETO-ACYL-COA THIOLASE 1, PKT2, peroxisomal 3-keto-acyl-CoA thiolase 2 |
-0.254 |
0.132 |
| 390 |
260242_at |
AT1G63650
|
AtEGL3, ATMYC-2, EGL1, EGL3, ENHANCER OF GLABRA 3 |
-0.253 |
0.212 |
| 391 |
253382_at |
AT4G33040
|
[Thioredoxin superfamily protein] |
-0.253 |
0.054 |
| 392 |
248853_at |
AT5G46570
|
BSK2, brassinosteroid-signaling kinase 2 |
-0.252 |
0.066 |
| 393 |
256781_at |
AT3G13650
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.252 |
-0.032 |
| 394 |
267010_at |
AT2G39250
|
SNZ, SCHNARCHZAPFEN |
-0.252 |
0.151 |
| 395 |
264444_at |
AT1G27360
|
SPL11, squamosa promoter-like 11 |
-0.251 |
0.112 |
| 396 |
251714_at |
AT3G56370
|
[Leucine-rich repeat protein kinase family protein] |
-0.251 |
0.054 |
| 397 |
254532_at |
AT4G19660
|
ATNPR4, NPR4, NPR1-like protein 4 |
-0.251 |
-0.169 |
| 398 |
260491_at |
AT1G51440
|
DALL2, DAD1-Like Lipase 2 |
-0.251 |
0.180 |
| 399 |
249355_at |
AT5G40500
|
unknown |
-0.249 |
0.152 |
| 400 |
257474_at |
AT1G80850
|
[DNA glycosylase superfamily protein] |
-0.249 |
-0.015 |
| 401 |
263883_at |
AT2G21830
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.249 |
0.087 |
| 402 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
-0.247 |
0.410 |
| 403 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
-0.246 |
0.369 |
| 404 |
248921_at |
AT5G45950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.246 |
0.057 |
| 405 |
264360_at |
AT1G03310
|
AtBE2, ATISA2, ARABIDOPSIS THALIANA ISOAMYLASE 2, BE2, BRANCHING ENZYME 2, DBE1, debranching enzyme 1, ISA2 |
-0.246 |
0.124 |
| 406 |
261661_at |
AT1G18360
|
[alpha/beta-Hydrolases superfamily protein] |
-0.245 |
0.171 |
| 407 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.245 |
0.080 |
| 408 |
265545_at |
AT2G28250
|
NCRK |
-0.244 |
0.062 |
| 409 |
250907_at |
AT5G03670
|
TRM28, TON1 Recruiting Motif 28 |
-0.242 |
0.162 |
| 410 |
252429_at |
AT3G47500
|
CDF3, cycling DOF factor 3 |
-0.242 |
0.306 |
| 411 |
254069_at |
AT4G25434
|
ATNUDT10, nudix hydrolase homolog 10, NUDT10, nudix hydrolase homolog 10 |
-0.242 |
0.215 |
| 412 |
253097_at |
AT4G37320
|
CYP81D5, cytochrome P450, family 81, subfamily D, polypeptide 5 |
-0.241 |
-0.075 |
| 413 |
266124_at |
AT2G45080
|
cycp3;1, cyclin p3;1 |
-0.240 |
0.138 |
| 414 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
-0.239 |
0.096 |
| 415 |
266479_at |
AT2G31160
|
LSH3, LIGHT SENSITIVE HYPOCOTYLS 3, OBO1, ORGAN BOUNDARY 1 |
-0.238 |
0.068 |
| 416 |
264314_at |
AT1G70420
|
unknown |
-0.237 |
0.195 |
| 417 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
-0.237 |
0.161 |
| 418 |
250955_at |
AT5G03190
|
CPUORF47, conserved peptide upstream open reading frame 47 |
-0.236 |
0.246 |
| 419 |
262891_at |
AT1G79460
|
ATKS, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE, ATKS1, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE 1, GA2, GA REQUIRING 2, KS, KS1, ENT-KAURENE SYNTHASE 1 |
-0.235 |
0.054 |
| 420 |
251024_at |
AT5G02180
|
[Transmembrane amino acid transporter family protein] |
-0.235 |
0.277 |
| 421 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
-0.235 |
-0.214 |
| 422 |
253145_at |
AT4G35560
|
DAW1, DUO1-activated WD40 1 |
-0.233 |
0.070 |
| 423 |
250558_at |
AT5G07990
|
CYP75B1, CYTOCHROME P450 75B1, D501, TT7, TRANSPARENT TESTA 7 |
-0.233 |
0.084 |
| 424 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
-0.232 |
-0.134 |
| 425 |
256652_at |
AT3G18850
|
LPAT5, lysophosphatidyl acyltransferase 5 |
-0.232 |
0.027 |
| 426 |
248232_at |
AT5G53760
|
ATMLO11, MILDEW RESISTANCE LOCUS O 11, MLO11, MILDEW RESISTANCE LOCUS O 11 |
-0.232 |
0.092 |
| 427 |
253886_at |
AT4G27710
|
CYP709B3, cytochrome P450, family 709, subfamily B, polypeptide 3 |
-0.231 |
0.072 |
| 428 |
259736_at |
AT1G64390
|
AtGH9C2, glycosyl hydrolase 9C2, GH9C2, glycosyl hydrolase 9C2 |
-0.230 |
0.220 |
| 429 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
-0.230 |
0.450 |
| 430 |
246591_at |
AT5G14880
|
KUP8, potassium uptake 8 |
-0.230 |
0.165 |
| 431 |
248596_at |
AT5G49330
|
ATMYB111, ARABIDOPSIS MYB DOMAIN PROTEIN 111, MYB111, myb domain protein 111, PFG3, PRODUCTION OF FLAVONOL GLYCOSIDES 3 |
-0.229 |
-0.162 |
| 432 |
245341_at |
AT4G16447
|
unknown |
-0.229 |
-0.057 |
| 433 |
259275_at |
AT3G01060
|
unknown |
-0.228 |
0.215 |
| 434 |
253302_at |
AT4G33660
|
unknown |
-0.228 |
0.015 |
| 435 |
256336_at |
AT1G72030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.228 |
0.191 |
| 436 |
260669_at |
AT1G19340
|
[Methyltransferase MT-A70 family protein] |
-0.227 |
-0.146 |
| 437 |
264978_at |
AT1G27120
|
[Galactosyltransferase family protein] |
-0.227 |
0.084 |
| 438 |
259081_at |
AT3G05030
|
ATNHX2, NHX2, sodium hydrogen exchanger 2 |
-0.226 |
0.167 |
| 439 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
-0.225 |
0.027 |
| 440 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.225 |
0.230 |
| 441 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.223 |
0.286 |
| 442 |
247918_at |
AT5G57610
|
[Protein kinase superfamily protein with octicosapeptide/Phox/Bem1p domain] |
-0.222 |
0.272 |
| 443 |
247577_at |
AT5G61290
|
[Flavin-binding monooxygenase family protein] |
-0.222 |
0.089 |
| 444 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-0.221 |
-0.035 |
| 445 |
254133_at |
AT4G24810
|
[Protein kinase superfamily protein] |
-0.219 |
-0.021 |
| 446 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.219 |
0.141 |
| 447 |
251174_at |
AT3G63200
|
PLA IIIB, PLP9, PATATIN-like protein 9 |
-0.217 |
0.069 |
| 448 |
250408_at |
AT5G10930
|
CIPK5, CBL-interacting protein kinase 5, SnRK3.24, SNF1-RELATED PROTEIN KINASE 3.24 |
-0.217 |
0.254 |
| 449 |
256319_at |
AT1G35910
|
TPPD, trehalose-6-phosphate phosphatase D |
-0.217 |
0.038 |
| 450 |
266357_at |
AT2G32290
|
BAM6, beta-amylase 6, BMY5, BETA-AMYLASE 5 |
-0.216 |
-0.048 |
| 451 |
250971_at |
AT5G02810
|
APRR7, PRR7, pseudo-response regulator 7 |
-0.216 |
0.158 |
| 452 |
254693_at |
AT4G17880
|
MYC4 |
-0.216 |
0.238 |
| 453 |
249303_at |
AT5G41460
|
unknown |
-0.215 |
-0.065 |
| 454 |
251725_at |
AT3G56260
|
unknown |
-0.214 |
0.114 |
| 455 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-0.214 |
0.143 |
| 456 |
254119_at |
AT4G24780
|
[Pectin lyase-like superfamily protein] |
-0.214 |
0.153 |
| 457 |
251952_at |
AT3G53650
|
[Histone superfamily protein] |
-0.214 |
-0.118 |
| 458 |
251658_at |
AT3G57020
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.213 |
0.174 |
| 459 |
246966_at |
AT5G24850
|
CRY3, cryptochrome 3 |
-0.213 |
0.197 |
| 460 |
263556_at |
AT2G16365
|
[F-box family protein] |
-0.213 |
0.052 |
| 461 |
247776_at |
AT5G58700
|
ATPLC4, PLC4, phosphatidylinositol-speciwc phospholipase C4 |
-0.212 |
0.146 |
| 462 |
266672_at |
AT2G29650
|
ANTR1, anion transporter 1, PHT4;1, phosphate transporter 4;1 |
-0.212 |
0.087 |
| 463 |
252619_at |
AT3G45210
|
unknown |
-0.212 |
0.118 |
| 464 |
262748_at |
AT1G28610
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.211 |
0.058 |
| 465 |
265894_at |
AT2G15050
|
LTP, lipid transfer protein, LTP7, lipid transfer protein 7 |
-0.211 |
0.013 |