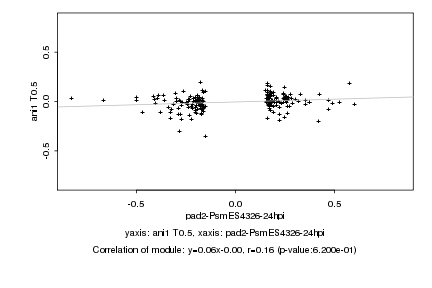
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
pad2-PsmES4326-24hpi |
Log2 signal ratio
ani1 T0.5 |
| 1 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.599 |
-0.021 |
| 2 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.575 |
0.183 |
| 3 |
262608_at |
AT1G14120
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.521 |
-0.003 |
| 4 |
257798_at |
AT3G15950
|
NAI2 |
0.486 |
-0.011 |
| 5 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
0.469 |
-0.073 |
| 6 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.465 |
0.013 |
| 7 |
258033_at |
AT3G21250
|
ABCC8, ATP-binding cassette C8, ATMRP6, ARABIDOPSIS THALIANA MULTIDRUG RESISTANCE-ASSOCIATED PROTEIN 6, MRP6, multidrug resistance-associated protein 6 |
0.423 |
0.080 |
| 8 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
0.419 |
-0.197 |
| 9 |
261266_at |
AT1G26770
|
AT-EXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXPA10, expansin A10, ATHEXP ALPHA 1.1, ARABIDOPSIS THALIANA EXPANSIN ALPHA 1.1, EXP10, EXPANSIN 10, EXPA10, expansin A10 |
0.370 |
-0.005 |
| 10 |
263904_at |
AT2G36380
|
ABCG34, ATP-binding cassette G34, ATPDR6, PLEIOTROPIC DRUG RESISTANCE 6, PDR6, pleiotropic drug resistance 6 |
0.369 |
-0.005 |
| 11 |
258369_at |
AT3G14310
|
ATPME3, pectin methylesterase 3, OZS2, OVERLY ZINC SENSITIVE 2, PME3, pectin methylesterase 3 |
0.352 |
-0.021 |
| 12 |
257895_at |
AT3G16950
|
LPD1, lipoamide dehydrogenase 1, ptlpd1 |
0.349 |
0.012 |
| 13 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
0.328 |
0.079 |
| 14 |
247218_at |
AT5G65010
|
ASN2, asparagine synthetase 2 |
0.314 |
0.001 |
| 15 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
0.300 |
0.023 |
| 16 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
0.284 |
-0.019 |
| 17 |
254034_at |
AT4G25960
|
ABCB2, ATP-binding cassette B2, PGP2, P-glycoprotein 2 |
0.280 |
0.036 |
| 18 |
263846_at |
AT2G36990
|
ATSIG6, SIGMA FACTOR 6, SIG6, SIGMA FACTOR 6, SIGF, RNApolymerase sigma-subunit F, SOLDAT8 |
0.274 |
0.074 |
| 19 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
0.272 |
-0.049 |
| 20 |
265076_at |
AT1G55490
|
CPN60B, chaperonin 60 beta, Cpn60beta1, chaperonin-60beta1, LEN1, LESION INITIATION 1 |
0.267 |
0.047 |
| 21 |
246371_at |
AT1G51940
|
LYK3, LysM-containing receptor-like kinase 3 |
0.260 |
-0.117 |
| 22 |
266035_at |
AT2G05990
|
ENR1, ENOYL-ACP REDUCTASE 1, MOD1, MOSAIC DEATH 1 |
0.260 |
0.027 |
| 23 |
263709_at |
AT1G09310
|
unknown |
0.259 |
-0.048 |
| 24 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
0.258 |
0.020 |
| 25 |
254649_at |
AT4G18570
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.254 |
0.051 |
| 26 |
247799_at |
AT5G58840
|
[Subtilase family protein] |
0.254 |
-0.001 |
| 27 |
247946_at |
AT5G57180
|
CIA2, chloroplast import apparatus 2 |
0.251 |
0.035 |
| 28 |
257773_at |
AT3G29185
|
unknown |
0.250 |
0.061 |
| 29 |
252468_at |
AT3G46970
|
ATPHS2, Arabidopsis thaliana alpha-glucan phosphorylase 2, PHS2, alpha-glucan phosphorylase 2 |
0.248 |
0.050 |
| 30 |
249807_at |
AT5G23870
|
[Pectinacetylesterase family protein] |
0.248 |
0.018 |
| 31 |
267606_at |
AT2G26640
|
KCS11, 3-ketoacyl-CoA synthase 11 |
0.247 |
0.042 |
| 32 |
247450_at |
AT5G62350
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.244 |
0.022 |
| 33 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
0.244 |
0.145 |
| 34 |
264720_at |
AT1G70080
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.243 |
-0.156 |
| 35 |
248921_at |
AT5G45950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.241 |
0.080 |
| 36 |
245524_at |
AT4G15920
|
AtSWEET17, SWEET17 |
0.240 |
-0.001 |
| 37 |
263513_at |
AT2G12400
|
unknown |
0.239 |
0.032 |
| 38 |
254562_at |
AT4G19230
|
CYP707A1, cytochrome P450, family 707, subfamily A, polypeptide 1 |
0.239 |
0.090 |
| 39 |
252115_at |
AT3G51600
|
LTP5, lipid transfer protein 5 |
0.230 |
-0.015 |
| 40 |
257206_at |
AT3G16530
|
[Legume lectin family protein] |
0.222 |
-0.129 |
| 41 |
257717_at |
AT3G18390
|
EMB1865, embryo defective 1865 |
0.220 |
-0.055 |
| 42 |
249026_at |
AT5G44785
|
OSB3, organellar single-stranded DNA binding protein 3 |
0.219 |
-0.004 |
| 43 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
0.219 |
-0.191 |
| 44 |
262598_at |
AT1G15260
|
unknown |
0.216 |
0.003 |
| 45 |
259302_at |
AT3G05120
|
ATGID1A, GA INSENSITIVE DWARF1A, GID1A, GA INSENSITIVE DWARF1A |
0.207 |
0.042 |
| 46 |
255829_at |
AT2G40540
|
ATKT2, ATKUP2, KT2, potassium transporter 2, KUP2, SHY3, TRK2 |
0.206 |
-0.039 |
| 47 |
254132_at |
AT4G24660
|
ATHB22, HOMEOBOX PROTEIN 22, HB22, homeobox protein 22, MEE68, MATERNAL EFFECT EMBRYO ARREST 68, ZHD2, ZINC FINGER HOMEODOMAIN 2 |
0.205 |
0.038 |
| 48 |
255403_at |
AT4G03400
|
DFL2, DWARF IN LIGHT 2, GH3-10 |
0.204 |
-0.009 |
| 49 |
253533_at |
AT4G31590
|
ATCSLC05, CELLULOSE-SYNTHASE LIKE C5, ATCSLC5, Cellulose-synthase-like C5, CSLC05, CELLULOSE-SYNTHASE LIKE C5, CSLC5, Cellulose-synthase-like C5 |
0.196 |
-0.008 |
| 50 |
261165_at |
AT1G34430
|
EMB3003, embryo defective 3003 |
0.194 |
0.011 |
| 51 |
249876_at |
AT5G23060
|
CaS, calcium sensing receptor |
0.192 |
0.066 |
| 52 |
257875_at |
AT3G17120
|
unknown |
0.190 |
-0.043 |
| 53 |
246268_at |
AT1G31800
|
CYP97A3, cytochrome P450, family 97, subfamily A, polypeptide 3, LUT5, LUTEIN DEFICIENT 5 |
0.189 |
0.101 |
| 54 |
246235_at |
AT4G36830
|
HOS3-1 |
0.188 |
-0.106 |
| 55 |
256836_at |
AT3G22960
|
PKP-ALPHA, PKP1, PLASTIDIAL PYRUVATE KINASE 1 |
0.185 |
0.006 |
| 56 |
247884_at |
AT5G57800
|
CER3, ECERIFERUM 3, FLP1, FACELESS POLLEN 1, WAX2, YRE |
0.182 |
0.016 |
| 57 |
250752_at |
AT5G05690
|
CBB3, CABBAGE 3, CPD, CONSTITUTIVE PHOTOMORPHOGENIC DWARF, CYP90, CYP90A, CYP90A1, CYTOCHROME P450 90A1, DWF3, DWARF 3 |
0.181 |
0.019 |
| 58 |
252032_at |
AT3G52150
|
PSRP2, plastid-specific ribosomal protein 2 |
0.178 |
0.086 |
| 59 |
257938_at |
AT3G19820
|
CBB1, CABBAGE 1, DIM, DIMINUTIA, DIM1, DIMINUTO 1, DWF1, DWARF 1, EVE1, ENHANCED VERY-LOW-FLUENCE RESPONSES 1 |
0.178 |
-0.003 |
| 60 |
262809_at |
AT1G11720
|
ATSS3, starch synthase 3, SS3, starch synthase 3 |
0.178 |
-0.036 |
| 61 |
262230_at |
AT1G68560
|
ATXYL1, ALPHA-XYLOSIDASE 1, AXY3, altered xyloglucan 3, TRG1, thermoinhibition resistant germination 1, XYL1, alpha-xylosidase 1 |
0.178 |
0.004 |
| 62 |
257172_at |
AT3G23700
|
[Nucleic acid-binding proteins superfamily] |
0.176 |
0.156 |
| 63 |
254181_at |
AT4G23940
|
ARC1, ACCUMULATION AND REPLICATION OF CHLOROPLASTS 1, FtsHi1, FTSH inactive protease 1 |
0.176 |
0.058 |
| 64 |
249614_at |
AT5G37300
|
WSD1 |
0.176 |
-0.037 |
| 65 |
251317_at |
AT3G61490
|
[Pectin lyase-like superfamily protein] |
0.175 |
0.086 |
| 66 |
248344_at |
AT5G52280
|
[Myosin heavy chain-related protein] |
0.173 |
0.064 |
| 67 |
247331_at |
AT5G63530
|
ATFP3, ARABIDOPSIS THALIANA FARNESYLATED PROTEIN 3, FP3, farnesylated protein 3 |
0.173 |
-0.082 |
| 68 |
253421_at |
AT4G32340
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.173 |
-0.089 |
| 69 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.173 |
-0.017 |
| 70 |
251036_at |
AT5G02160
|
unknown |
0.172 |
0.109 |
| 71 |
263053_at |
AT2G13440
|
[glucose-inhibited division family A protein] |
0.171 |
-0.012 |
| 72 |
260106_at |
AT1G35420
|
[alpha/beta-Hydrolases superfamily protein] |
0.170 |
0.079 |
| 73 |
258076_at |
AT3G25920
|
RPL15, ribosomal protein L15 |
0.170 |
0.012 |
| 74 |
251519_at |
AT3G59400
|
GUN4, GENOMES UNCOUPLED 4 |
0.170 |
0.018 |
| 75 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.169 |
-0.066 |
| 76 |
267088_at |
AT2G38140
|
PSRP4, plastid-specific ribosomal protein 4 |
0.169 |
-0.012 |
| 77 |
265414_at |
AT2G16660
|
[Major facilitator superfamily protein] |
0.169 |
-0.047 |
| 78 |
261190_at |
AT1G32990
|
PRPL11, plastid ribosomal protein l11 |
0.169 |
-0.002 |
| 79 |
254221_at |
AT4G23820
|
[Pectin lyase-like superfamily protein] |
0.169 |
-0.004 |
| 80 |
256277_at |
AT3G12120
|
AtFAD2, FAD2, fatty acid desaturase 2 |
0.167 |
0.034 |
| 81 |
266421_at |
AT2G38540
|
ATLTP1, ARABIDOPSIS THALIANA LIPID TRANSFER PROTEIN 1, LP1, lipid transfer protein 1, LTP1, LIPID TRANSFER PROTEIN 1 |
0.167 |
0.038 |
| 82 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
0.166 |
-0.024 |
| 83 |
264986_at |
AT1G27130
|
ATGSTU13, glutathione S-transferase tau 13, GST12, GLUTATHIONE S-TRANSFERASE 12, GSTU13, glutathione S-transferase tau 13 |
0.166 |
-0.039 |
| 84 |
261661_at |
AT1G18360
|
[alpha/beta-Hydrolases superfamily protein] |
0.164 |
0.051 |
| 85 |
248235_at |
AT5G53860
|
emb2737, embryo defective 2737, EMB64, EMBRYO DEFECTIVE 64 |
0.164 |
0.014 |
| 86 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.161 |
-0.165 |
| 87 |
249742_at |
AT5G24490
|
[30S ribosomal protein, putative] |
0.161 |
0.069 |
| 88 |
267635_at |
AT2G42220
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.160 |
0.166 |
| 89 |
252116_at |
AT3G51510
|
unknown |
0.159 |
0.187 |
| 90 |
264057_at |
AT2G28550
|
RAP2.7, related to AP2.7, TOE1, TARGET OF EARLY ACTIVATION TAGGED (EAT) 1 |
0.159 |
0.072 |
| 91 |
258432_at |
AT3G16570
|
ATRALF23, ARABIDOPSIS RAPID ALKALINIZATION FACTOR 23, RALF23, rapid alkalinization factor 23 |
0.159 |
0.096 |
| 92 |
247040_at |
AT5G67150
|
[HXXXD-type acyl-transferase family protein] |
0.159 |
0.031 |
| 93 |
247862_at |
AT5G58250
|
EMB3143, EMBRYO DEFECTIVE 3143 |
0.158 |
0.006 |
| 94 |
256805_at |
AT3G20930
|
ORRM1, Organelle RRM protein 1 |
0.157 |
0.115 |
| 95 |
262693_at |
AT1G62780
|
unknown |
0.157 |
0.073 |
| 96 |
259848_at |
AT1G72180
|
[Leucine-rich receptor-like protein kinase family protein] |
0.156 |
0.004 |
| 97 |
250245_at |
AT5G13690
|
CYL1, CYCLOPS 1, NAGLU, N-ACETYL-GLUCOSAMINIDASE |
0.156 |
-0.003 |
| 98 |
250884_at |
AT5G03940
|
54CP, 54 CHLOROPLAST PROTEIN, CPSRP54, chloroplast signal recognition particle 54 kDa subunit, FFC, FIFTY-FOUR CHLOROPLAST HOMOLOGUE, SRP54CP, SIGNAL RECOGNITION PARTICLE 54 KDA SUBUNIT CHLOROPLAST PROTEIN |
0.154 |
0.033 |
| 99 |
249035_at |
AT5G44190
|
ATGLK2, GLK2, GOLDEN2-like 2, GPRI2, GBF'S PRO-RICH REGION-INTERACTING FACTOR 2 |
0.151 |
0.117 |
| 100 |
260713_at |
AT1G17615
|
[Disease resistance protein (TIR-NBS class)] |
-0.831 |
0.032 |
| 101 |
248918_at |
AT5G45890
|
AtSAG12, SAG12, senescence-associated gene 12 |
-0.667 |
0.013 |
| 102 |
266900_at |
AT2G34610
|
unknown |
-0.502 |
0.017 |
| 103 |
254277_at |
AT4G22680
|
AtMYB85, myb domain protein 85, MYB85, myb domain protein 85 |
-0.501 |
0.041 |
| 104 |
249562_at |
AT5G38350
|
[Disease resistance protein (NBS-LRR class) family] |
-0.473 |
-0.107 |
| 105 |
247324_at |
AT5G64190
|
unknown |
-0.414 |
0.052 |
| 106 |
257950_at |
AT3G21780
|
UGT71B6, UDP-glucosyl transferase 71B6 |
-0.413 |
0.030 |
| 107 |
266571_at |
AT2G23830
|
[PapD-like superfamily protein] |
-0.406 |
-0.013 |
| 108 |
256176_at |
AT1G51640
|
ATEXO70G2, exocyst subunit exo70 family protein G2, EXO70G2, exocyst subunit exo70 family protein G2 |
-0.396 |
0.038 |
| 109 |
255074_at |
AT4G09100
|
[RING/U-box superfamily protein] |
-0.393 |
0.069 |
| 110 |
252448_at |
AT3G47050
|
[Glycosyl hydrolase family protein] |
-0.380 |
-0.111 |
| 111 |
265582_at |
AT2G20030
|
[RING/U-box superfamily protein] |
-0.364 |
0.066 |
| 112 |
253837_at |
AT4G27850
|
[Glycine-rich protein family] |
-0.359 |
0.011 |
| 113 |
248846_at |
AT5G46500
|
unknown |
-0.340 |
-0.057 |
| 114 |
252447_at |
AT3G47040
|
[Glycosyl hydrolase family protein] |
-0.329 |
-0.110 |
| 115 |
245257_at |
AT4G14640
|
AtCML8, calmodulin-like 8, CAM8, calmodulin 8, CML8 |
-0.328 |
-0.172 |
| 116 |
266780_at |
AT2G29110
|
ATGLR2.8, glutamate receptor 2.8, GLR2.8, GLR2.8, glutamate receptor 2.8 |
-0.326 |
-0.074 |
| 117 |
260278_at |
AT1G80590
|
ATWRKY66, WRKY66, WRKY DNA-binding protein 66 |
-0.315 |
-0.028 |
| 118 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.304 |
0.085 |
| 119 |
247178_at |
AT5G65205
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.298 |
0.004 |
| 120 |
255341_at |
AT4G04500
|
CRK37, cysteine-rich RLK (RECEPTOR-like protein kinase) 37 |
-0.298 |
0.039 |
| 121 |
260561_at |
AT2G43580
|
[Chitinase family protein] |
-0.292 |
-0.130 |
| 122 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.288 |
-0.067 |
| 123 |
251456_at |
AT3G60120
|
BGLU27, beta glucosidase 27 |
-0.286 |
-0.296 |
| 124 |
245160_at |
AT2G33080
|
AtRLP28, receptor like protein 28, RLP28, receptor like protein 28 |
-0.285 |
0.020 |
| 125 |
261241_at |
AT1G32950
|
[Subtilase family protein] |
-0.282 |
0.003 |
| 126 |
247753_at |
AT5G59070
|
[UDP-Glycosyltransferase superfamily protein] |
-0.279 |
-0.124 |
| 127 |
247105_at |
AT5G66630
|
DAR5, DA1-related protein 5 |
-0.275 |
-0.175 |
| 128 |
252572_at |
AT3G45290
|
ATMLO3, MILDEW RESISTANCE LOCUS O 3, MLO3, MILDEW RESISTANCE LOCUS O 3 |
-0.273 |
-0.032 |
| 129 |
252681_at |
AT3G44350
|
anac061, NAC domain containing protein 61, NAC061, NAC domain containing protein 61 |
-0.271 |
-0.009 |
| 130 |
262827_at |
AT1G13100
|
CYP71B29, cytochrome P450, family 71, subfamily B, polypeptide 29 |
-0.264 |
0.105 |
| 131 |
259893_at |
AT1G71390
|
AtRLP11, receptor like protein 11, RLP11, receptor like protein 11 |
-0.250 |
-0.004 |
| 132 |
246498_at |
AT5G16230
|
[Plant stearoyl-acyl-carrier-protein desaturase family protein] |
-0.250 |
-0.020 |
| 133 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
-0.242 |
-0.025 |
| 134 |
249055_at |
AT5G44460
|
CML43, calmodulin like 43 |
-0.240 |
-0.055 |
| 135 |
261366_at |
AT1G53100
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.240 |
0.030 |
| 136 |
264537_at |
AT1G55610
|
BRL1, BRI1 like |
-0.239 |
0.001 |
| 137 |
259251_at |
AT3G07600
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.234 |
-0.132 |
| 138 |
255546_at |
AT4G01910
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.232 |
0.028 |
| 139 |
245677_at |
AT1G56660
|
unknown |
-0.231 |
0.055 |
| 140 |
260005_at |
AT1G67920
|
unknown |
-0.223 |
-0.176 |
| 141 |
245611_at |
AT4G14390
|
[Ankyrin repeat family protein] |
-0.223 |
-0.043 |
| 142 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
-0.218 |
-0.063 |
| 143 |
263658_at |
AT1G04490
|
unknown |
-0.218 |
-0.028 |
| 144 |
250049_at |
AT5G17780
|
[alpha/beta-Hydrolases superfamily protein] |
-0.214 |
0.036 |
| 145 |
265659_at |
AT2G25440
|
AtRLP20, receptor like protein 20, RLP20, receptor like protein 20 |
-0.213 |
0.007 |
| 146 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-0.206 |
0.041 |
| 147 |
259065_at |
AT3G07520
|
ATGLR1.4, GLR1.4, glutamate receptor 1.4 |
-0.205 |
0.017 |
| 148 |
249312_at |
AT5G41550
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.205 |
-0.103 |
| 149 |
247435_at |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
-0.204 |
-0.084 |
| 150 |
259320_at |
AT3G01080
|
ATWRKY58, WRKY DNA-BINDING PROTEIN 58, WRKY58, WRKY DNA-binding protein 58 |
-0.203 |
-0.051 |
| 151 |
267372_at |
AT2G26290
|
ARSK1, root-specific kinase 1 |
-0.200 |
-0.115 |
| 152 |
260840_at |
AT1G29050
|
TBL38, TRICHOME BIREFRINGENCE-LIKE 38 |
-0.198 |
-0.034 |
| 153 |
246373_at |
AT1G51860
|
[Leucine-rich repeat protein kinase family protein] |
-0.198 |
-0.007 |
| 154 |
254253_at |
AT4G23320
|
CRK24, cysteine-rich RLK (RECEPTOR-like protein kinase) 24 |
-0.197 |
-0.031 |
| 155 |
265586_at |
AT2G19990
|
PR-1-LIKE, pathogenesis-related protein-1-like |
-0.196 |
0.025 |
| 156 |
260908_at |
AT1G02580
|
EMB173, EMBRYO DEFECTIVE 173, FIS1, FERTILIZATION INDEPENDENT SEED 1, MEA, MEDEA, SDG5, SET DOMAIN-CONTAINING PROTEIN 5 |
-0.196 |
-0.003 |
| 157 |
264082_at |
AT2G28570
|
unknown |
-0.195 |
-0.077 |
| 158 |
252346_at |
AT3G48650
|
[pseudogene] |
-0.194 |
-0.008 |
| 159 |
249705_at |
AT5G35580
|
[Protein kinase superfamily protein] |
-0.193 |
-0.005 |
| 160 |
256464_at |
AT1G32560
|
AtLEA4-1, Late Embryogenesis Abundant 4-1, LEA4-1, Late Embryogenesis Abundant 4-1 |
-0.192 |
0.069 |
| 161 |
259766_at |
AT1G64360
|
unknown |
-0.191 |
0.027 |
| 162 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
-0.191 |
-0.002 |
| 163 |
248133_at |
AT5G54850
|
unknown |
-0.190 |
-0.018 |
| 164 |
263655_at |
AT1G04500
|
[CCT motif family protein] |
-0.189 |
0.034 |
| 165 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
-0.186 |
-0.017 |
| 166 |
262254_at |
AT1G53920
|
GLIP5, GDSL-motif lipase 5 |
-0.185 |
0.020 |
| 167 |
264685_at |
AT1G65610
|
ATGH9A2, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9A2, KOR2, KORRIGAN 2 |
-0.185 |
-0.032 |
| 168 |
254905_at |
AT4G11170
|
RMG1, Resistance Methylated Gene 1 |
-0.184 |
-0.001 |
| 169 |
256778_at |
AT3G13782
|
NAP1;4, nucleosome assembly protein1;4, NFA04, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP A 04, NFA4, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP A 4 |
-0.183 |
0.050 |
| 170 |
250024_at |
AT5G18270
|
ANAC087, Arabidopsis NAC domain containing protein 87 |
-0.183 |
0.027 |
| 171 |
261058_at |
AT1G01370
|
CENH3, CENTROMERIC HISTONE H3, HTR12 |
-0.182 |
0.048 |
| 172 |
259809_at |
AT1G49800
|
unknown |
-0.182 |
-0.004 |
| 173 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
-0.181 |
0.196 |
| 174 |
251277_at |
AT3G61760
|
ADL1B, DYNAMIN-like 1B, DL1B, DYNAMIN-like 1B |
-0.180 |
-0.016 |
| 175 |
253737_at |
AT4G28703
|
[RmlC-like cupins superfamily protein] |
-0.178 |
-0.049 |
| 176 |
252300_at |
AT3G49160
|
[pyruvate kinase family protein] |
-0.177 |
-0.069 |
| 177 |
261429_at |
AT1G18860
|
ATWRKY61, WRKY DNA-BINDING PROTEIN 61, WRKY61, WRKY DNA-binding protein 61 |
-0.176 |
-0.112 |
| 178 |
258942_at |
AT3G09960
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.175 |
-0.129 |
| 179 |
259036_at |
AT3G09220
|
LAC7, laccase 7 |
-0.174 |
-0.076 |
| 180 |
248416_at |
AT5G51630
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.171 |
-0.063 |
| 181 |
251076_at |
AT5G01970
|
unknown |
-0.170 |
0.037 |
| 182 |
254796_at |
AT4G13000
|
[AGC (cAMP-dependent, cGMP-dependent and protein kinase C) kinase family protein] |
-0.170 |
-0.021 |
| 183 |
266902_at |
AT2G34580
|
unknown |
-0.170 |
0.004 |
| 184 |
259472_at |
AT1G18910
|
[zinc ion binding] |
-0.169 |
-0.043 |
| 185 |
253806_at |
AT4G28270
|
ATRMA2, RMA2, RING membrane-anchor 2 |
-0.168 |
0.118 |
| 186 |
266596_at |
AT2G46150
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.168 |
-0.071 |
| 187 |
250064_at |
AT5G17890
|
CHS3, CHILLING SENSITIVE 3, DAR4, DA1-related protein 4 |
-0.168 |
0.012 |
| 188 |
247448_at |
AT5G62770
|
unknown |
-0.167 |
-0.038 |
| 189 |
260734_at |
AT1G17600
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.166 |
0.010 |
| 190 |
267418_at |
AT2G35000
|
ATL9, Arabidopsis toxicos en levadura 9 |
-0.166 |
-0.052 |
| 191 |
259534_at |
AT1G12290
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.166 |
-0.098 |
| 192 |
255306_at |
AT4G04740
|
ATCPK23, CPK23, calcium-dependent protein kinase 23 |
-0.163 |
0.096 |
| 193 |
259635_at |
AT1G56360
|
ATPAP6, PAP6, purple acid phosphatase 6 |
-0.162 |
-0.049 |
| 194 |
248726_at |
AT5G47960
|
ATRABA4C, RAB GTPase homolog A4C, RABA4C, RAB GTPase homolog A4C, SMG1, SMALL MOLECULAR WEIGHT G-PROTEIN 1 |
-0.162 |
-0.066 |
| 195 |
248700_at |
AT5G48400
|
ATGLR1.2, GLR1.2, GLUTAMATE RECEPTOR 1.2 |
-0.156 |
-0.050 |
| 196 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
-0.156 |
0.109 |
| 197 |
256499_at |
AT1G36640
|
unknown |
-0.156 |
-0.353 |