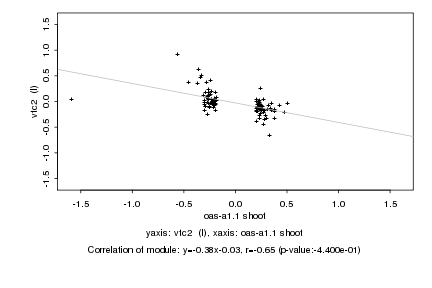
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
oas-a1.1 shoot |
Log2 signal ratio
vtc2 (I) |
| 1 |
257670_at |
AT3G20340
|
unknown |
0.499 |
-0.027 |
| 2 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
0.472 |
-0.213 |
| 3 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.418 |
-0.059 |
| 4 |
249144_at |
AT5G43270
|
SPL2, squamosa promoter binding protein-like 2 |
0.374 |
-0.182 |
| 5 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
0.373 |
-0.312 |
| 6 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.370 |
-0.155 |
| 7 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
0.348 |
-0.032 |
| 8 |
258033_at |
AT3G21250
|
ABCC8, ATP-binding cassette C8, ATMRP6, ARABIDOPSIS THALIANA MULTIDRUG RESISTANCE-ASSOCIATED PROTEIN 6, MRP6, multidrug resistance-associated protein 6 |
0.344 |
-0.174 |
| 9 |
262231_at |
AT1G68740
|
PHO1;H1 |
0.336 |
-0.118 |
| 10 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.321 |
-0.651 |
| 11 |
259729_at |
AT1G77640
|
[Integrase-type DNA-binding superfamily protein] |
0.313 |
-0.069 |
| 12 |
257076_at |
AT3G19680
|
unknown |
0.303 |
-0.152 |
| 13 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.292 |
-0.321 |
| 14 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
0.282 |
-0.259 |
| 15 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
0.278 |
-0.348 |
| 16 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
0.276 |
-0.174 |
| 17 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
0.268 |
-0.173 |
| 18 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
0.266 |
-0.431 |
| 19 |
265339_at |
AT2G18230
|
AtPPa2, pyrophosphorylase 2, PPa2, pyrophosphorylase 2 |
0.264 |
-0.211 |
| 20 |
246996_at |
AT5G67420
|
ASL39, ASYMMETRIC LEAVES2-LIKE 39, LBD37, LOB domain-containing protein 37 |
0.263 |
0.045 |
| 21 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
0.261 |
-0.095 |
| 22 |
251734_at |
AT3G56270
|
unknown |
0.257 |
-0.125 |
| 23 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
0.250 |
-0.123 |
| 24 |
260427_at |
AT1G72430
|
SAUR78, SMALL AUXIN UPREGULATED RNA 78 |
0.247 |
-0.144 |
| 25 |
258932_at |
AT3G10150
|
ATPAP16, PAP16, purple acid phosphatase 16 |
0.246 |
-0.161 |
| 26 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
0.244 |
-0.075 |
| 27 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
0.243 |
-0.223 |
| 28 |
258621_at |
AT3G02830
|
PNT1, PENTA 1, ZFN1, zinc finger protein 1 |
0.242 |
-0.124 |
| 29 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.240 |
0.254 |
| 30 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
0.234 |
-0.066 |
| 31 |
256603_at |
AT3G28270
|
unknown |
0.231 |
-0.325 |
| 32 |
261819_at |
AT1G11410
|
[S-locus lectin protein kinase family protein] |
0.229 |
-0.155 |
| 33 |
247617_at |
AT5G60270
|
LecRK-I.7, L-type lectin receptor kinase I.7 |
0.229 |
-0.062 |
| 34 |
262577_at |
AT1G15290
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.227 |
-0.118 |
| 35 |
246926_at |
AT5G25240
|
unknown |
0.226 |
-0.148 |
| 36 |
249016_at |
AT5G44750
|
ATREV1, ARABIDOPSIS THALIANA HOMOLOG OF REVERSIONLESS 1, REV1 |
0.225 |
-0.083 |
| 37 |
251735_at |
AT3G56090
|
ATFER3, ferritin 3, FER3, ferritin 3 |
0.224 |
-0.004 |
| 38 |
265678_at |
AT2G31970
|
ATRAD50, RAD50 |
0.224 |
0.038 |
| 39 |
246075_at |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
0.224 |
-0.268 |
| 40 |
253166_at |
AT4G35290
|
ATGLR3.2, ATGLUR2, GLR3.2, GLUTAMATE RECEPTOR 3.2, GLUR2, glutamate receptor 2 |
0.222 |
-0.123 |
| 41 |
264700_at |
AT1G70100
|
unknown |
0.219 |
-0.026 |
| 42 |
252667_at |
AT3G44200
|
ATNEK6, NIMA-RELATED KINASE6, IBO1, NEK6, NIMA (never in mitosis, gene A)-related 6 |
0.218 |
-0.084 |
| 43 |
266464_at |
AT2G47800
|
ABCC4, ATP-binding cassette C4, ATMRP4, multidrug resistance-associated protein 4, EST3, MRP4, multidrug resistance-associated protein 4 |
0.217 |
0.002 |
| 44 |
261117_at |
AT1G75310
|
AUL1, auxilin-like 1 |
0.217 |
-0.109 |
| 45 |
245084_at |
AT2G23290
|
AtMYB70, myb domain protein 70, MYB70, myb domain protein 70 |
0.212 |
-0.101 |
| 46 |
259399_at |
AT1G17710
|
AtPEPC1, Arabidopsis thaliana phosphoethanolamine/phosphocholine phosphatase 1, PEPC1, phosphoethanolamine/phosphocholine phosphatase 1 |
0.211 |
-0.046 |
| 47 |
250050_at |
AT5G17790
|
VAR3, VARIEGATED 3 |
0.209 |
-0.080 |
| 48 |
255488_at |
AT4G02630
|
[Protein kinase superfamily protein] |
0.209 |
-0.188 |
| 49 |
258470_at |
AT3G06035
|
[Glycoprotein membrane precursor GPI-anchored] |
0.206 |
-0.101 |
| 50 |
251309_at |
AT3G61220
|
SDR1, short-chain dehydrogenase/reductase 1 |
0.206 |
-0.085 |
| 51 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
0.202 |
-0.179 |
| 52 |
267319_at |
AT2G34660
|
ABCC2, ATP-binding cassette C2, AtABCC2, Arabidopsis thaliana ATP-binding cassette C2, ATMRP2, multidrug resistance-associated protein 2, EST4, MRP2, multidrug resistance-associated protein 2 |
0.202 |
-0.163 |
| 53 |
263483_at |
AT2G04030
|
AtHsp90.5, HEAT SHOCK PROTEIN 90.5, AtHsp90C, CR88, EMB1956, EMBRYO DEFECTIVE 1956, Hsp88.1, HEAT SHOCK PROTEIN 88.1, HSP90.5, HEAT SHOCK PROTEIN 90.5 |
0.202 |
0.019 |
| 54 |
250429_at |
AT5G10470
|
KAC1, kinesin like protein for actin based chloroplast movement 1, KCA1, KINESIN CDKA;1 ASSOCIATED 1 |
0.201 |
-0.135 |
| 55 |
256302_at |
AT1G69526
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.198 |
-0.373 |
| 56 |
254662_at |
AT4G18270
|
ATTRANS11, ARABIDOPSIS THALIANA TRANSLOCASE 11, TRANS11, translocase 11 |
0.195 |
0.056 |
| 57 |
245286_at |
AT4G14880
|
ATCYS-3A, CYTACS1, OASA1, O-acetylserine (thiol) lyase (OAS-TL) isoform A1, OLD3, ONSET OF LEAF DEATH 3 |
-1.597 |
0.040 |
| 58 |
251673_at |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
-0.566 |
0.934 |
| 59 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
-0.461 |
0.389 |
| 60 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
-0.370 |
0.363 |
| 61 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
-0.363 |
0.640 |
| 62 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
-0.344 |
0.484 |
| 63 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
-0.331 |
0.507 |
| 64 |
252533_at |
AT3G46110
|
[LOCATED IN: plasma membrane] |
-0.313 |
0.130 |
| 65 |
255856_at |
AT1G66940
|
[protein kinase-related] |
-0.309 |
0.025 |
| 66 |
266489_at |
AT2G35190
|
ATNPSN11, NPSN11, novel plant snare 11, NSPN11 |
-0.307 |
-0.018 |
| 67 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
-0.307 |
-0.164 |
| 68 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.305 |
-0.048 |
| 69 |
261032_at |
AT1G17430
|
[alpha/beta-Hydrolases superfamily protein] |
-0.300 |
0.190 |
| 70 |
262813_at |
AT1G11670
|
[MATE efflux family protein] |
-0.297 |
-0.084 |
| 71 |
258764_at |
AT3G10720
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.295 |
-0.084 |
| 72 |
259143_at |
AT3G10190
|
[Calcium-binding EF-hand family protein] |
-0.285 |
0.087 |
| 73 |
251232_at |
AT3G62780
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.281 |
0.375 |
| 74 |
257053_at |
AT3G15210
|
ATERF-4, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 4, ATERF4, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 4, ERF4, ethylene responsive element binding factor 4, RAP2.5, RELATED TO AP2 5 |
-0.277 |
-0.018 |
| 75 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
-0.277 |
-0.246 |
| 76 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.275 |
0.106 |
| 77 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
-0.270 |
0.130 |
| 78 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.268 |
0.250 |
| 79 |
262236_at |
AT1G48330
|
unknown |
-0.267 |
0.190 |
| 80 |
260688_at |
AT1G17665
|
unknown |
-0.263 |
-0.026 |
| 81 |
262698_at |
AT1G75960
|
[AMP-dependent synthetase and ligase family protein] |
-0.262 |
0.074 |
| 82 |
246562_at |
AT5G15580
|
LNG1, LONGIFOLIA1, TRM2, TON1 Recruiting Motif 2 |
-0.262 |
0.046 |
| 83 |
248775_at |
AT5G47850
|
CCR4, CRINKLY4 related 4 |
-0.256 |
0.120 |
| 84 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
-0.254 |
-0.092 |
| 85 |
261252_at |
AT1G05810
|
ARA, ARA-1, ATRAB11D, ATRABA5E, ARABIDOPSIS THALIANA RAB GTPASE HOMOLOG A5E, RABA5E, RAB GTPase homolog A5E |
-0.252 |
-0.103 |
| 86 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
-0.250 |
-0.021 |
| 87 |
249242_at |
AT5G42250
|
[Zinc-binding alcohol dehydrogenase family protein] |
-0.250 |
0.145 |
| 88 |
264055_at |
AT2G28590
|
[Protein kinase superfamily protein] |
-0.247 |
-0.035 |
| 89 |
258787_at |
AT3G11840
|
PUB24, plant U-box 24 |
-0.246 |
0.425 |
| 90 |
245784_at |
AT1G32190
|
[alpha/beta-Hydrolases superfamily protein] |
-0.241 |
0.046 |
| 91 |
257701_at |
AT3G12710
|
[DNA glycosylase superfamily protein] |
-0.239 |
0.050 |
| 92 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
-0.233 |
0.198 |
| 93 |
256069_at |
AT1G13740
|
AFP2, ABI five binding protein 2 |
-0.233 |
-0.033 |
| 94 |
254349_at |
AT4G22250
|
[RING/U-box superfamily protein] |
-0.229 |
-0.016 |
| 95 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.224 |
-0.040 |
| 96 |
252661_at |
AT3G44450
|
unknown |
-0.223 |
0.015 |
| 97 |
267040_at |
AT2G34300
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.220 |
-0.114 |
| 98 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
-0.216 |
-0.006 |
| 99 |
247910_at |
AT5G57410
|
[Afadin/alpha-actinin-binding protein] |
-0.214 |
0.038 |
| 100 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
-0.212 |
-0.062 |
| 101 |
246952_at |
AT5G04820
|
ATOFP13, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 13, OFP13, ovate family protein 13 |
-0.210 |
0.057 |
| 102 |
267289_at |
AT2G23770
|
LYK4, LysM-containing receptor-like kinase 4 |
-0.209 |
0.039 |
| 103 |
248855_at |
AT5G46590
|
anac096, NAC domain containing protein 96, NAC096, NAC domain containing protein 96 |
-0.207 |
0.066 |
| 104 |
249783_at |
AT5G24270
|
ATSOS3, SALT OVERLY SENSITIVE 3, CBL4, CALCINEURIN B-LIKE PROTEIN 4, SOS3, SALT OVERLY SENSITIVE 3 |
-0.201 |
0.034 |
| 105 |
260944_at |
AT1G45130
|
AtBGAL5, BGAL5, beta-galactosidase 5 |
-0.201 |
-0.040 |
| 106 |
260009_at |
AT1G67950
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.197 |
-0.028 |
| 107 |
260676_at |
AT1G19450
|
[Major facilitator superfamily protein] |
-0.196 |
-0.159 |
| 108 |
258207_at |
AT3G14050
|
AT-RSH2, RELA-SPOT HOMOLOG 2, ATRSH2, RELA/SPOT HOMOLOG 2, RSH2, RELA/SPOT homolog 2 |
-0.196 |
0.180 |
| 109 |
247183_at |
AT5G65440
|
unknown |
-0.194 |
0.025 |
| 110 |
252378_at |
AT3G47570
|
[Leucine-rich repeat protein kinase family protein] |
-0.192 |
0.085 |
| 111 |
252834_at |
AT4G40070
|
[RING/U-box superfamily protein] |
-0.192 |
0.021 |