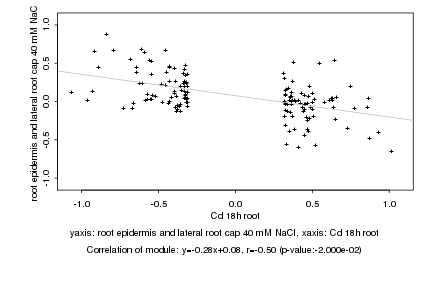
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Cd 18h root |
Log2 signal ratio
root epidermis and lateral root cap 40 mM NaCl |
| 1 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
1.011 |
-0.645 |
| 2 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
0.924 |
-0.399 |
| 3 |
251869_at |
AT3G54500
|
LNK2, night light-inducible and clock-regulated 2 |
0.869 |
-0.470 |
| 4 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
0.860 |
0.040 |
| 5 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
0.858 |
-0.074 |
| 6 |
261684_at |
AT1G47400
|
unknown |
0.770 |
-0.091 |
| 7 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.746 |
0.200 |
| 8 |
247323_at |
AT5G64170
|
LNK1, night light-inducible and clock-regulated 1 |
0.723 |
-0.344 |
| 9 |
264635_at |
AT1G65500
|
unknown |
0.652 |
0.053 |
| 10 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
0.648 |
-0.223 |
| 11 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.643 |
0.547 |
| 12 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
0.637 |
-0.071 |
| 13 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
0.629 |
0.016 |
| 14 |
262838_at |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.626 |
0.044 |
| 15 |
256319_at |
AT1G35910
|
TPPD, trehalose-6-phosphate phosphatase D |
0.621 |
0.043 |
| 16 |
246000_at |
AT5G20820
|
SAUR76, SMALL AUXIN UPREGULATED RNA 76 |
0.610 |
0.026 |
| 17 |
256131_at |
AT1G13600
|
AtbZIP58, basic leucine-zipper 58, bZIP58, basic leucine-zipper 58 |
0.578 |
-0.008 |
| 18 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
0.546 |
0.499 |
| 19 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.518 |
-0.569 |
| 20 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
0.514 |
0.034 |
| 21 |
265569_at |
AT2G05620
|
AtPGR5, PGR5, proton gradient regulation 5 |
0.503 |
-0.183 |
| 22 |
246229_at |
AT4G37160
|
sks15, SKU5 similar 15 |
0.497 |
-0.102 |
| 23 |
246967_at |
AT5G24860
|
ATFPF1, ARABIDOPSIS FLOWERING PROMOTING FACTOR 1, FPF1, FLOWERING PROMOTING FACTOR 1 |
0.496 |
-0.012 |
| 24 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
0.495 |
0.111 |
| 25 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.486 |
-0.068 |
| 26 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
0.481 |
-0.219 |
| 27 |
252958_at |
AT4G38620
|
ATMYB4, myb domain protein 4, MYB4, myb domain protein 4 |
0.476 |
0.197 |
| 28 |
250243_at |
AT5G13630
|
ABAR, ABA-BINDING PROTEIN, CCH, CONDITIONAL CHLORINA, CCH1, CHLH, H SUBUNIT OF MG-CHELATASE, GUN5, GENOMES UNCOUPLED 5 |
0.473 |
0.075 |
| 29 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
0.472 |
-0.382 |
| 30 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
0.467 |
-0.243 |
| 31 |
251727_at |
AT3G56290
|
unknown |
0.466 |
-0.357 |
| 32 |
254907_at |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.466 |
-0.013 |
| 33 |
247232_at |
AT5G64940
|
ATATH13, ARABIDOPSIS THALIANA ABC2 HOMOLOG 13, ATH13, ABC2 homolog 13, ATOSA1, A. THALIANA OXIDATIVE STRESS-RELATED ABC1-LIKE PROTEIN 1, OSA1, OXIDATIVE STRESS-RELATED ABC1-LIKE PROTEIN 1 |
0.458 |
-0.198 |
| 34 |
252929_at |
AT4G38970
|
AtFBA2, FBA2, fructose-bisphosphate aldolase 2 |
0.453 |
-0.034 |
| 35 |
256352_at |
AT1G54970
|
ATPRP1, proline-rich protein 1, PRP1, proline-rich protein 1, RHS7, ROOT HAIR SPECIFIC 7 |
0.449 |
-0.434 |
| 36 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
0.449 |
0.080 |
| 37 |
248356_at |
AT5G52350
|
ATEXO70A3, exocyst subunit exo70 family protein A3, EXO70A3, exocyst subunit exo70 family protein A3 |
0.448 |
-0.026 |
| 38 |
266672_at |
AT2G29650
|
ANTR1, anion transporter 1, PHT4;1, phosphate transporter 4;1 |
0.445 |
-0.104 |
| 39 |
261881_at |
AT1G80760
|
NIP6, NIP6;1, NOD26-like intrinsic protein 6;1, NLM7 |
0.440 |
-0.121 |
| 40 |
255812_at |
AT4G10310
|
ATHKT1, HKT1, high-affinity K+ transporter 1, HKT1;1 |
0.430 |
-0.070 |
| 41 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
0.429 |
0.117 |
| 42 |
256880_at |
AT3G26450
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.420 |
-0.019 |
| 43 |
262608_at |
AT1G14120
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.410 |
0.018 |
| 44 |
247914_at |
AT5G57540
|
AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 |
0.410 |
-0.595 |
| 45 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
0.390 |
0.002 |
| 46 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
0.384 |
0.014 |
| 47 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.381 |
-0.362 |
| 48 |
257650_at |
AT3G16800
|
[Protein phosphatase 2C family protein] |
0.373 |
0.522 |
| 49 |
266828_at |
AT2G22930
|
[UDP-Glycosyltransferase superfamily protein] |
0.367 |
0.016 |
| 50 |
251060_at |
AT5G01820
|
ATCIPK14, ATSR1, serine/threonine protein kinase 1, CIPK14, CBL-INTERACTING PROTEIN KINASE 14, PKS24, SOS2-like protein kinase 24, SnRK3.15, SNF1-RELATED PROTEIN KINASE 3.15, SR1, serine/threonine protein kinase 1 |
0.366 |
0.269 |
| 51 |
255011_at |
AT4G10040
|
CYTC-2, cytochrome c-2 |
0.365 |
-0.183 |
| 52 |
266108_at |
AT2G37900
|
[Major facilitator superfamily protein] |
0.362 |
-0.026 |
| 53 |
262526_at |
AT1G17050
|
AtSPS2, SPS2, solanesyl diphosphate synthase 2 |
0.361 |
0.076 |
| 54 |
260014_at |
AT1G68010
|
ATHPR1, HPR, hydroxypyruvate reductase |
0.361 |
0.003 |
| 55 |
257129_at |
AT3G20100
|
CYP705A19, cytochrome P450, family 705, subfamily A, polypeptide 19 |
0.359 |
0.118 |
| 56 |
245558_at |
AT4G15430
|
[ERD (early-responsive to dehydration stress) family protein] |
0.357 |
0.065 |
| 57 |
256681_at |
AT3G52340
|
ATSPP2, SUCROSE-PHOSPHATASE 2, SPP2, sucrose-6F-phosphate phosphohydrolase 2 |
0.356 |
0.072 |
| 58 |
262743_at |
AT1G29020
|
[Calcium-binding EF-hand family protein] |
0.354 |
-0.138 |
| 59 |
252594_at |
AT3G45680
|
[Major facilitator superfamily protein] |
0.351 |
0.023 |
| 60 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
0.351 |
-0.381 |
| 61 |
252202_at |
AT3G50300
|
[HXXXD-type acyl-transferase family protein] |
0.340 |
0.175 |
| 62 |
259537_at |
AT1G12370
|
PHR1, photolyase 1, UVR2, UV RESISTANCE 2 |
0.338 |
-0.030 |
| 63 |
252429_at |
AT3G47500
|
CDF3, cycling DOF factor 3 |
0.337 |
-0.128 |
| 64 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
0.327 |
-0.554 |
| 65 |
261690_at |
AT1G50090
|
AtBCAT7, BCAT7, branched-chain amino acid transaminase 7 |
0.327 |
0.163 |
| 66 |
247452_at |
AT5G62430
|
CDF1, cycling DOF factor 1 |
0.325 |
0.087 |
| 67 |
260481_at |
AT1G10960
|
ATFD1, ferredoxin 1, FD1, ferredoxin 1 |
0.324 |
-0.112 |
| 68 |
255105_at |
AT4G08620
|
SULTR1;1, sulphate transporter 1;1 |
0.323 |
-0.020 |
| 69 |
260025_at |
AT1G30070
|
[SGS domain-containing protein] |
0.322 |
-0.302 |
| 70 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
0.322 |
0.151 |
| 71 |
261133_at |
AT1G19715
|
[Mannose-binding lectin superfamily protein] |
0.321 |
0.102 |
| 72 |
257252_at |
AT3G24170
|
ATGR1, glutathione-disulfide reductase, GR1, glutathione-disulfide reductase |
0.320 |
-0.033 |
| 73 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.317 |
0.304 |
| 74 |
248619_at |
AT5G49630
|
AAP6, amino acid permease 6 |
0.317 |
0.003 |
| 75 |
255814_at |
AT1G19900
|
[glyoxal oxidase-related protein] |
0.315 |
-0.193 |
| 76 |
259150_at |
AT3G10320
|
[Glycosyltransferase family 61 protein] |
0.307 |
0.366 |
| 77 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-1.071 |
0.130 |
| 78 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
-0.963 |
0.023 |
| 79 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
-0.932 |
0.143 |
| 80 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
-0.917 |
0.656 |
| 81 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
-0.894 |
0.451 |
| 82 |
253322_at |
AT4G33980
|
unknown |
-0.843 |
0.883 |
| 83 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
-0.797 |
0.667 |
| 84 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
-0.732 |
-0.090 |
| 85 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
-0.687 |
0.559 |
| 86 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
-0.672 |
-0.085 |
| 87 |
264774_at |
AT1G22890
|
unknown |
-0.665 |
-0.023 |
| 88 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
-0.650 |
0.451 |
| 89 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
-0.647 |
0.386 |
| 90 |
267382_at |
AT2G44300
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.630 |
0.238 |
| 91 |
262452_at |
AT1G11210
|
unknown |
-0.614 |
0.686 |
| 92 |
245546_at |
AT4G15290
|
ATCSLB05, ATCSLB5, CELLULOSE SYNTHASE LIKE 5, CSLB05 |
-0.606 |
0.244 |
| 93 |
261613_at |
AT1G49720
|
ABF1, abscisic acid responsive element-binding factor 1, AtABF1 |
-0.594 |
0.652 |
| 94 |
252924_at |
AT4G39070
|
BBX20, B-box domain protein 20, BZS1, BZS1 |
-0.588 |
0.025 |
| 95 |
267254_at |
AT2G23030
|
SNRK2-9, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-9, SNRK2.9, SNF1-related protein kinase 2.9 |
-0.577 |
0.033 |
| 96 |
256746_at |
AT3G29320
|
PHS1, alpha-glucan phosphorylase 1 |
-0.572 |
0.104 |
| 97 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
-0.565 |
0.547 |
| 98 |
254001_at |
AT4G26260
|
MIOX4, myo-inositol oxygenase 4 |
-0.553 |
0.039 |
| 99 |
253309_at |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
-0.551 |
0.361 |
| 100 |
257670_at |
AT3G20340
|
unknown |
-0.549 |
0.523 |
| 101 |
246703_at |
AT5G28080
|
WNK9 |
-0.546 |
0.033 |
| 102 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
-0.542 |
0.085 |
| 103 |
264425_at |
AT1G61750
|
[Receptor-like protein kinase-related family protein] |
-0.520 |
0.068 |
| 104 |
263556_at |
AT2G16365
|
[F-box family protein] |
-0.486 |
0.228 |
| 105 |
259316_at |
AT3G01175
|
unknown |
-0.480 |
-0.008 |
| 106 |
259595_at |
AT1G28050
|
BBX13, B-box domain protein 13 |
-0.461 |
0.221 |
| 107 |
266983_at |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
-0.457 |
0.668 |
| 108 |
248148_at |
AT5G54930
|
[AT hook motif-containing protein] |
-0.453 |
0.388 |
| 109 |
250194_at |
AT5G14550
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.438 |
-0.018 |
| 110 |
247582_at |
AT5G60760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.434 |
0.005 |
| 111 |
259992_at |
AT1G67970
|
AT-HSFA8, HSFA8, heat shock transcription factor A8 |
-0.431 |
0.464 |
| 112 |
247525_at |
AT5G61380
|
APRR1, AtTOC1, TIMING OF CAB EXPRESSION 1, PRR1, PSEUDO-RESPONSE REGULATOR 1, TOC1, TIMING OF CAB EXPRESSION 1 |
-0.431 |
0.262 |
| 113 |
249742_at |
AT5G24490
|
[30S ribosomal protein, putative] |
-0.431 |
0.456 |
| 114 |
251996_at |
AT3G52840
|
BGAL2, beta-galactosidase 2 |
-0.417 |
0.054 |
| 115 |
252357_at |
AT3G48410
|
[alpha/beta-Hydrolases superfamily protein] |
-0.401 |
0.117 |
| 116 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
-0.399 |
0.439 |
| 117 |
264045_at |
AT2G22450
|
AtRIBA2, RIBA2, homolog of ribA 2 |
-0.398 |
0.143 |
| 118 |
248382_at |
AT5G51890
|
[Peroxidase superfamily protein] |
-0.394 |
-0.067 |
| 119 |
260412_at |
AT1G69830
|
AMY3, alpha-amylase-like 3, ATAMY3, ALPHA-AMYLASE-LIKE 3 |
-0.392 |
0.265 |
| 120 |
263919_at |
AT2G36470
|
unknown |
-0.386 |
-0.127 |
| 121 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
-0.384 |
0.077 |
| 122 |
265560_at |
AT2G05520
|
ATGRP-3, GLYCINE-RICH PROTEIN 3, ATGRP3, ARABIDOPSIS GLYCINE RICH PROTEIN 3, GRP-3, glycine-rich protein 3, GRP3, GLYCINE-RICH PROTEIN 3 |
-0.380 |
-0.050 |
| 123 |
264597_at |
AT1G04620
|
HCAR, 7-hydroxymethyl chlorophyll a (HMChl) reductase |
-0.379 |
-0.094 |
| 124 |
264551_at |
AT1G09460
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.366 |
-0.064 |
| 125 |
251132_at |
AT5G01200
|
[Duplicated homeodomain-like superfamily protein] |
-0.362 |
0.199 |
| 126 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
-0.362 |
-0.029 |
| 127 |
245353_at |
AT4G16000
|
unknown |
-0.362 |
-0.130 |
| 128 |
259363_at |
AT1G13270
|
MAP1B, METHIONINE AMINOPEPTIDASE 1B, MAP1C, methionine aminopeptidase 1B |
-0.352 |
0.146 |
| 129 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
-0.344 |
0.368 |
| 130 |
253614_at |
AT4G30350
|
SMXL2, SMAX1-like 2 |
-0.342 |
0.241 |
| 131 |
250724_at |
AT5G06330
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.337 |
0.051 |
| 132 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.336 |
0.425 |
| 133 |
248969_at |
AT5G45310
|
unknown |
-0.335 |
0.135 |
| 134 |
247013_at |
AT5G67480
|
ATBT4, BT4, BTB and TAZ domain protein 4 |
-0.333 |
0.206 |
| 135 |
252468_at |
AT3G46970
|
ATPHS2, Arabidopsis thaliana alpha-glucan phosphorylase 2, PHS2, alpha-glucan phosphorylase 2 |
-0.332 |
0.268 |
| 136 |
263737_at |
AT1G60010
|
unknown |
-0.328 |
0.063 |
| 137 |
255252_at |
AT4G04990
|
unknown |
-0.328 |
0.091 |
| 138 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
-0.326 |
0.352 |
| 139 |
264204_at |
AT1G22710
|
ATSUC2, ARABIDOPSIS THALIANA SUCROSE-PROTON SYMPORTER 2, SUC2, sucrose-proton symporter 2, SUT1, SUCROSE TRANSPORTER 1 |
-0.326 |
0.479 |
| 140 |
257154_at |
AT3G27210
|
unknown |
-0.326 |
0.104 |
| 141 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
-0.324 |
0.028 |
| 142 |
265131_at |
AT1G23760
|
JP630, PG3, POLYGALACTURONASE 3 |
-0.323 |
0.242 |
| 143 |
260024_at |
AT1G30080
|
[Glycosyl hydrolase superfamily protein] |
-0.322 |
-0.001 |
| 144 |
255070_at |
AT4G09020
|
ATISA3, ISA3, isoamylase 3 |
-0.321 |
0.254 |
| 145 |
253237_at |
AT4G34240
|
ALDH3, aldehyde dehydrogenase 3, ALDH3I1, aldehyde dehydrogenase 3I1 |
-0.318 |
-0.052 |
| 146 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
-0.317 |
0.126 |
| 147 |
263548_at |
AT2G21660
|
ATGRP7, GLYCINE RICH PROTEIN 7, CCR2, cold, circadian rhythm, and rna binding 2, GR-RBP7, GLYCINE-RICH RNA-BINDING PROTEIN 7, GRP7, GLYCINE-RICH RNA-BINDING PROTEIN 7 |
-0.316 |
-0.012 |
| 148 |
266358_at |
AT2G32280
|
VCC, VASCULATURE COMPLEXITY AND CONNECTIVITY |
-0.315 |
0.046 |
| 149 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
-0.314 |
0.208 |
| 150 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
-0.314 |
0.363 |
| 151 |
247882_at |
AT5G57785
|
unknown |
-0.313 |
0.130 |