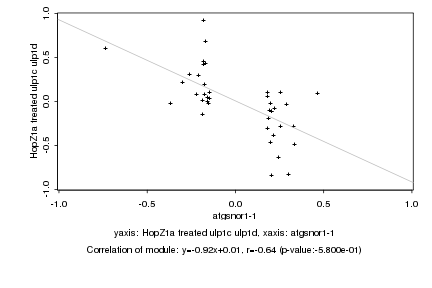
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
atgsnor1-1 |
Log2 signal ratio
HopZ1a treated ulp1c ulp1d |
| 1 |
252014_at |
AT3G52870
|
[IQ calmodulin-binding motif family protein] |
0.463 |
0.096 |
| 2 |
264698_at |
AT1G70200
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.331 |
-0.483 |
| 3 |
250327_at |
AT5G12050
|
unknown |
0.326 |
-0.282 |
| 4 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
0.297 |
-0.822 |
| 5 |
261924_at |
AT1G22550
|
[Major facilitator superfamily protein] |
0.284 |
-0.029 |
| 6 |
262164_at |
AT1G78070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.255 |
0.113 |
| 7 |
262245_at |
AT1G48240
|
ATNPSN12, NPSN12, novel plant snare 12 |
0.250 |
-0.283 |
| 8 |
265918_at |
AT2G15090
|
KCS8, 3-ketoacyl-CoA synthase 8 |
0.242 |
-0.636 |
| 9 |
250418_at |
AT5G11240
|
[transducin family protein / WD-40 repeat family protein] |
0.217 |
-0.069 |
| 10 |
267578_at |
AT2G30695
|
unknown |
0.215 |
-0.386 |
| 11 |
250104_at |
AT5G16610
|
unknown |
0.201 |
-0.108 |
| 12 |
259700_at |
AT1G68980
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.201 |
-0.112 |
| 13 |
256237_at |
AT3G12610
|
DRT100, DNA-DAMAGE REPAIR/TOLERATION 100 |
0.200 |
-0.840 |
| 14 |
252911_at |
AT4G39510
|
CYP96A12, cytochrome P450, family 96, subfamily A, polypeptide 12 |
0.196 |
-0.022 |
| 15 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
0.195 |
-0.465 |
| 16 |
247875_at |
AT5G57720
|
[AP2/B3-like transcriptional factor family protein] |
0.188 |
-0.101 |
| 17 |
245031_at |
AT2G26360
|
[Mitochondrial substrate carrier family protein] |
0.187 |
-0.190 |
| 18 |
251584_at |
AT3G58620
|
TTL4, tetratricopetide-repeat thioredoxin-like 4 |
0.181 |
0.061 |
| 19 |
254150_at |
AT4G24350
|
[Phosphorylase superfamily protein] |
0.180 |
0.114 |
| 20 |
257300_at |
AT3G28080
|
UMAMIT47, Usually multiple acids move in and out Transporters 47 |
0.176 |
-0.305 |
| 21 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
-0.737 |
0.605 |
| 22 |
267016_at |
AT2G39160
|
unknown |
-0.373 |
-0.015 |
| 23 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
-0.305 |
0.226 |
| 24 |
247064_at |
AT5G66890
|
[Leucine-rich repeat (LRR) family protein] |
-0.265 |
0.314 |
| 25 |
253123_at |
AT4G36000
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.225 |
0.085 |
| 26 |
245755_at |
AT1G35210
|
unknown |
-0.213 |
0.305 |
| 27 |
245891_at |
AT5G09220
|
AAP2, amino acid permease 2 |
-0.189 |
-0.141 |
| 28 |
267227_at |
AT2G44030
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.189 |
0.015 |
| 29 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
-0.186 |
0.429 |
| 30 |
265853_at |
AT2G42360
|
[RING/U-box superfamily protein] |
-0.185 |
0.933 |
| 31 |
255723_at |
AT3G29575
|
AFP3, ABI five binding protein 3 |
-0.185 |
0.457 |
| 32 |
262140_at |
AT1G52470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.180 |
0.080 |
| 33 |
252488_at |
AT3G46700
|
[UDP-Glycosyltransferase superfamily protein] |
-0.178 |
0.197 |
| 34 |
253181_at |
AT4G35180
|
LHT7, LYS/HIS transporter 7 |
-0.175 |
0.694 |
| 35 |
260243_at |
AT1G63720
|
unknown |
-0.172 |
0.437 |
| 36 |
265068_at |
AT1G55550
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.162 |
0.048 |
| 37 |
260381_at |
AT1G73860
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.160 |
0.009 |
| 38 |
256500_at |
AT1G36620
|
[copia-like retrotransposon family, has a 3.6e-54 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
-0.158 |
-0.014 |
| 39 |
266381_at |
AT2G14670
|
ATSUC8, sucrose-proton symporter 8, SUC8, sucrose-proton symporter 8 |
-0.152 |
0.108 |
| 40 |
246904_at |
AT5G25480
|
AtDNMT2, DNA METHYLTRANSFERASE 2, DNMT2, DNA methyltransferase-2 |
-0.151 |
0.037 |