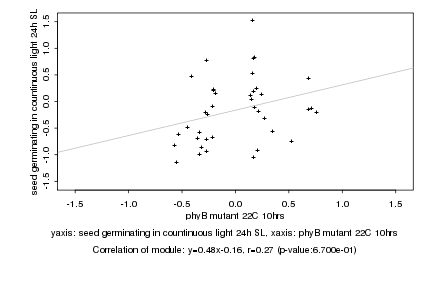
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
phyB mutant 22C 10hrs |
Log2 signal ratio
seed germinating in countinuous light 24h SL |
| 1 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
0.750 |
-0.202 |
| 2 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
0.705 |
-0.122 |
| 3 |
254452_at |
AT4G21100
|
DDB1B, damaged DNA binding protein 1B |
0.680 |
-0.149 |
| 4 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
0.678 |
0.440 |
| 5 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
0.521 |
-0.734 |
| 6 |
261425_at |
AT1G18880
|
AtNPF2.9, NPF2.9, NRT1/ PTR family 2.9, NRT1.9, nitrate transporter 1.9 |
0.339 |
-0.555 |
| 7 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
0.269 |
-0.309 |
| 8 |
254451_at |
AT4G21090
|
ATMFDX2, ARABIDOPSIS MITOCHONDRIAL FERREDOXIN 2, MFDX2, MITOCHONDRIAL FERREDOXIN 2 |
0.240 |
0.148 |
| 9 |
251368_at |
AT3G61380
|
TRM14, TON1 Recruiting Motif 14 |
0.209 |
-0.177 |
| 10 |
259149_at |
AT3G10340
|
PAL4, phenylalanine ammonia-lyase 4 |
0.199 |
-0.918 |
| 11 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
0.196 |
0.259 |
| 12 |
262646_at |
AT1G62800
|
ASP4, aspartate aminotransferase 4 |
0.178 |
-0.107 |
| 13 |
259403_at |
AT1G17745
|
PGDH, 3-phosphoglycerate dehydrogenase, PGDH2, phosphoglycerate dehydrogenase 2 |
0.176 |
0.846 |
| 14 |
266352_at |
AT2G01610
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.167 |
-1.036 |
| 15 |
250556_at |
AT5G07920
|
ATDGK1, DIACYLGLYCEROL KINASE 1, DGK1, diacylglycerol kinase1 |
0.166 |
0.190 |
| 16 |
257701_at |
AT3G12710
|
[DNA glycosylase superfamily protein] |
0.160 |
0.811 |
| 17 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
0.151 |
0.535 |
| 18 |
255080_at |
AT4G09030
|
AGP10, arabinogalactan protein 10, ATAGP10 |
0.151 |
1.541 |
| 19 |
255727_at |
AT1G25510
|
[Eukaryotic aspartyl protease family protein] |
0.148 |
0.055 |
| 20 |
261050_at |
AT1G01260
|
JAM2, Jasmonate Associated MYC2 LIKE 2 |
0.135 |
0.131 |
| 21 |
252345_at |
AT3G48640
|
unknown |
-0.576 |
-0.810 |
| 22 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
-0.555 |
-1.136 |
| 23 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.541 |
-0.605 |
| 24 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
-0.455 |
-0.472 |
| 25 |
263852_at |
AT2G04450
|
ATNUDT6, nudix hydrolase homolog 6, ATNUDX6, Arabidopsis thaliana nucleoside diphosphate linked to some moiety X 6, NUDT6, nudix hydrolase homolog 6, NUDX6, nucleoside diphosphates linked to some moiety X 6 |
-0.418 |
0.487 |
| 26 |
255503_at |
AT4G02420
|
LecRK-IV.4, L-type lectin receptor kinase IV.4 |
-0.360 |
-0.691 |
| 27 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
-0.338 |
-0.992 |
| 28 |
247602_at |
AT5G60900
|
RLK1, receptor-like protein kinase 1 |
-0.337 |
-0.569 |
| 29 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
-0.318 |
-0.853 |
| 30 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
-0.289 |
-0.194 |
| 31 |
253414_at |
AT4G33050
|
AtIQM1, EDA39, embryo sac development arrest 39, IQM1, IQ-motif protein 1 |
-0.280 |
-0.932 |
| 32 |
249783_at |
AT5G24270
|
ATSOS3, SALT OVERLY SENSITIVE 3, CBL4, CALCINEURIN B-LIKE PROTEIN 4, SOS3, SALT OVERLY SENSITIVE 3 |
-0.278 |
0.774 |
| 33 |
251621_at |
AT3G57700
|
[Protein kinase superfamily protein] |
-0.274 |
-0.696 |
| 34 |
254500_at |
AT4G20110
|
BP80-3;1, binding protein of 80 kDa 3;1, VSR3;1, VACUOLAR SORTING RECEPTOR 3;1, VSR7, VACUOLAR SORTING RECEPTOR 7 |
-0.264 |
-0.231 |
| 35 |
260211_at |
AT1G74440
|
unknown |
-0.216 |
-0.664 |
| 36 |
265161_at |
AT1G30900
|
BP80-3;3, binding protein of 80 kDa 3;3, VSR3;3, VACUOLAR SORTING RECEPTOR 3;3, VSR6, VACUOLAR SORTING RECEPTOR 6 |
-0.216 |
-0.083 |
| 37 |
264507_at |
AT1G09415
|
NIMIN-3, NIM1-interacting 3 |
-0.209 |
0.210 |
| 38 |
252037_at |
AT3G51920
|
ATCML9, CAM9, calmodulin 9, CML9, CALMODULIN LIKE PROTEIN 9 |
-0.208 |
0.244 |
| 39 |
258227_at |
AT3G15620
|
UVR3, UV REPAIR DEFECTIVE 3 |
-0.189 |
0.158 |