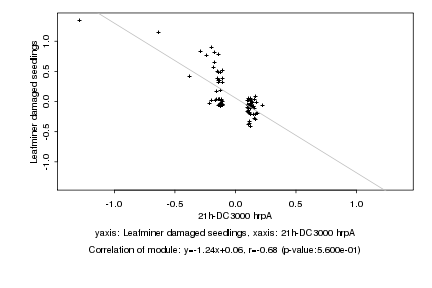
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
21h-DC3000 hrpA |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
258034_at |
AT3G21300
|
[RNA methyltransferase family protein] |
0.222 |
-0.060 |
| 2 |
256675_at |
AT3G52170
|
[DNA binding] |
0.180 |
-0.188 |
| 3 |
263039_at |
AT1G23280
|
[MAK16 protein-related] |
0.173 |
-0.011 |
| 4 |
266598_at |
AT2G46110
|
KPHMT1, ketopantoate hydroxymethyltransferase 1, PANB1 |
0.173 |
-0.187 |
| 5 |
247056_at |
AT5G66750
|
ATDDM1, CHA1, CHR01, CHROMATIN REMODELING 1, CHR1, chromatin remodeling 1, DDM1, DECREASED DNA METHYLATION 1, SOM1, SOMNIFEROUS 1, SOM4 |
0.164 |
-0.207 |
| 6 |
247197_at |
AT5G65240
|
[Leucine-rich repeat protein kinase family protein] |
0.162 |
0.095 |
| 7 |
254043_at |
AT4G25990
|
CIL |
0.161 |
-0.282 |
| 8 |
251256_at |
AT3G62300
|
unknown |
0.153 |
-0.278 |
| 9 |
266957_at |
AT2G34640
|
HMR, HEMERA, PTAC12, plastid transcriptionally active 12, TAC12 |
0.153 |
-0.114 |
| 10 |
256284_at |
AT3G12530
|
PSF2 |
0.151 |
0.044 |
| 11 |
248984_at |
AT5G45140
|
NRPC2, nuclear RNA polymerase C2 |
0.145 |
-0.070 |
| 12 |
258589_at |
AT3G04290
|
ATLTL1, LTL1, Li-tolerant lipase 1 |
0.143 |
-0.200 |
| 13 |
260826_at |
AT1G06710
|
MTSF1, mitochondrial stability factor 1 |
0.141 |
-0.077 |
| 14 |
256781_at |
AT3G13650
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.140 |
-0.001 |
| 15 |
263679_at |
AT1G59990
|
EMB3108, EMBRYO DEFECTIVE 3108, HS3, HEAVY SEED 3, RH22, RNA helicase 22 |
0.133 |
-0.076 |
| 16 |
252625_at |
AT3G44750
|
ATHD2A, HD2A, HISTONE DEACETYLASE 2A, HDA3, histone deacetylase 3, HDT1 |
0.130 |
0.002 |
| 17 |
266668_at |
AT2G29760
|
OTP81, ORGANELLE TRANSCRIPT PROCESSING 81 |
0.126 |
-0.066 |
| 18 |
251956_at |
AT3G53460
|
CP29, chloroplast RNA-binding protein 29 |
0.125 |
-0.099 |
| 19 |
251476_at |
AT3G59670
|
unknown |
0.124 |
-0.212 |
| 20 |
246562_at |
AT5G15580
|
LNG1, LONGIFOLIA1, TRM2, TON1 Recruiting Motif 2 |
0.123 |
-0.404 |
| 21 |
247032_at |
AT5G67240
|
SDN3, small RNA degrading nuclease 3 |
0.123 |
-0.026 |
| 22 |
256288_at |
AT3G12270
|
ATPRMT3, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 3, PRMT3, protein arginine methyltransferase 3 |
0.122 |
-0.048 |
| 23 |
265895_at |
AT2G15000
|
unknown |
0.120 |
0.030 |
| 24 |
253492_at |
AT4G31810
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
0.119 |
0.034 |
| 25 |
259172_at |
AT3G03630
|
CS26, cysteine synthase 26 |
0.118 |
-0.108 |
| 26 |
264560_at |
AT1G55820
|
unknown |
0.118 |
0.060 |
| 27 |
247643_at |
AT5G60450
|
ARF4, auxin response factor 4 |
0.114 |
-0.331 |
| 28 |
245174_at |
AT2G47500
|
[P-loop nucleoside triphosphate hydrolases superfamily protein with CH (Calponin Homology) domain] |
0.113 |
-0.363 |
| 29 |
247362_at |
AT5G63140
|
ATPAP29, purple acid phosphatase 29, PAP29, PAP29, purple acid phosphatase 29 |
0.112 |
-0.193 |
| 30 |
249826_at |
AT5G23310
|
FSD3, Fe superoxide dismutase 3 |
0.110 |
-0.171 |
| 31 |
256862_at |
AT3G23940
|
[dehydratase family] |
0.107 |
-0.133 |
| 32 |
259680_at |
AT1G77690
|
LAX3, like AUX1 3 |
0.107 |
-0.370 |
| 33 |
256027_at |
AT1G34160
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.103 |
0.022 |
| 34 |
246303_at |
AT3G51870
|
[Mitochondrial substrate carrier family protein] |
0.103 |
-0.111 |
| 35 |
251935_at |
AT3G54090
|
FLN1, fructokinase-like 1 |
0.103 |
-0.062 |
| 36 |
266612_at |
AT2G14860
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
0.101 |
-0.172 |
| 37 |
251833_at |
AT3G55160
|
unknown |
0.100 |
-0.050 |
| 38 |
259690_at |
AT1G63160
|
EMB2811, EMBRYO DEFECTIVE 2811, RFC2, replication factor C 2 |
0.100 |
0.055 |
| 39 |
258089_at |
AT3G14740
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.099 |
-0.096 |
| 40 |
260779_at |
AT1G14650
|
[SWAP (Suppressor-of-White-APricot)/surp domain-containing protein / ubiquitin family protein] |
0.098 |
-0.043 |
| 41 |
263435_at |
AT2G28600
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.098 |
0.018 |
| 42 |
266687_at |
AT2G19670
|
ATPRMT1A, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 1A, PRMT1A, protein arginine methyltransferase 1A |
0.097 |
-0.158 |
| 43 |
259286_at |
AT3G11480
|
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
-1.292 |
1.360 |
| 44 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
-0.641 |
1.152 |
| 45 |
258322_at |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
-0.384 |
0.427 |
| 46 |
254447_at |
AT4G20860
|
[FAD-binding Berberine family protein] |
-0.293 |
0.837 |
| 47 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
-0.244 |
0.767 |
| 48 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
-0.216 |
-0.032 |
| 49 |
250589_at |
AT5G07700
|
AtMYB76, myb domain protein 76, MYB76, myb domain protein 76 |
-0.205 |
0.033 |
| 50 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
-0.201 |
0.900 |
| 51 |
263032_at |
AT1G23850
|
unknown |
-0.189 |
0.580 |
| 52 |
247175_at |
AT5G65280
|
GCL1, GCR2-like 1 |
-0.179 |
0.649 |
| 53 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
-0.174 |
0.820 |
| 54 |
245640_at |
AT1G25330
|
CES, CESTA, HAF, HALF FILLED |
-0.171 |
0.017 |
| 55 |
250881_at |
AT5G04080
|
unknown |
-0.163 |
0.169 |
| 56 |
262416_at |
AT1G49390
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.160 |
0.047 |
| 57 |
264052_at |
AT2G22330
|
CYP79B3, cytochrome P450, family 79, subfamily B, polypeptide 3 |
-0.153 |
0.383 |
| 58 |
266661_at |
AT2G25820
|
ESE2, ethylene and salt inducible 2 |
-0.150 |
0.501 |
| 59 |
259548_at |
AT1G35260
|
MLP165, MLP-like protein 165 |
-0.147 |
-0.064 |
| 60 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
-0.146 |
0.355 |
| 61 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
-0.146 |
0.490 |
| 62 |
257154_at |
AT3G27210
|
unknown |
-0.143 |
0.793 |
| 63 |
246620_at |
AT5G36220
|
CYP81D1, cytochrome P450, family 81, subfamily D, polypeptide 1, CYP91A1, CYTOCHROME P450 91A1 |
-0.142 |
0.329 |
| 64 |
246333_at |
AT3G44840
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.140 |
0.044 |
| 65 |
248898_at |
AT5G46370
|
ATKCO2, CA2+ ACTIVATED OUTWARD RECTIFYING K+ CHANNEL 2, ATTPK2, TANDEM PORE K+ CHANNEL 2, KCO2, Ca2+ activated outward rectifying K+ channel 2 |
-0.139 |
-0.035 |
| 66 |
260461_at |
AT1G10980
|
[Lung seven transmembrane receptor family protein] |
-0.136 |
0.039 |
| 67 |
267035_at |
AT2G38400
|
AGT3, alanine:glyoxylate aminotransferase 3 |
-0.132 |
0.364 |
| 68 |
246876_at |
AT5G26140
|
ATLOG9, LOG9, LONELY GUY 9 |
-0.132 |
0.039 |
| 69 |
265802_at |
AT2G35733
|
unknown |
-0.127 |
-0.074 |
| 70 |
259483_at |
AT1G43980
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.126 |
-0.025 |
| 71 |
246197_at |
AT4G37010
|
CEN2, centrin 2 |
-0.125 |
0.491 |
| 72 |
250633_at |
AT5G07460
|
ATMSRA2, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE 2, PMSR2, peptidemethionine sulfoxide reductase 2 |
-0.124 |
0.197 |
| 73 |
264993_at |
AT1G67290
|
GLOX1, glyoxal oxidase 1 |
-0.120 |
-0.017 |
| 74 |
255804_at |
AT4G10220
|
unknown |
-0.119 |
0.034 |
| 75 |
251101_at |
AT5G01680
|
ATCHX26, ARABIDOPSIS THALIANA CATION/HYDROGEN EXCHANGER 26, CHX26, cation/H+ exchanger 26 |
-0.118 |
-0.042 |
| 76 |
248345_at |
AT5G52290
|
SHOC1, SHORTAGE IN CHIASMATA 1 |
-0.116 |
0.024 |
| 77 |
257364_at |
AT2G45940
|
unknown |
-0.113 |
0.316 |
| 78 |
256006_at |
AT1G34070
|
unknown |
-0.111 |
-0.044 |
| 79 |
263103_at |
AT2G05290
|
unknown |
-0.110 |
0.002 |
| 80 |
257450_at |
AT1G10530
|
unknown |
-0.109 |
0.519 |
| 81 |
249509_at |
AT5G38390
|
[F-box/RNI-like superfamily protein] |
-0.109 |
-0.060 |
| 82 |
259708_at |
AT1G77420
|
[alpha/beta-Hydrolases superfamily protein] |
-0.107 |
0.398 |
| 83 |
266723_at |
AT2G03190
|
ASK16, SKP1-like 16, SK16, SKP1-like 16 |
-0.107 |
-0.041 |