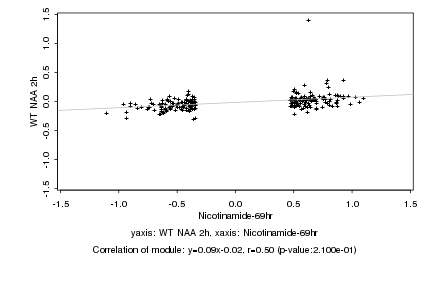
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Nicotinamide-69hr |
Log2 signal ratio
WT NAA 2h |
| 1 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
1.092 |
0.065 |
| 2 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
1.057 |
-0.012 |
| 3 |
252511_at |
AT3G46280
|
[protein kinase-related] |
1.029 |
0.071 |
| 4 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.984 |
-0.037 |
| 5 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
0.964 |
0.088 |
| 6 |
248703_at |
AT5G48430
|
[Eukaryotic aspartyl protease family protein] |
0.923 |
0.095 |
| 7 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.921 |
0.058 |
| 8 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
0.919 |
0.375 |
| 9 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
0.898 |
0.087 |
| 10 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
0.878 |
0.094 |
| 11 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.874 |
0.119 |
| 12 |
259120_at |
AT3G02240
|
GLV4, GOLVEN 4, RGF7, root meristem growth factor 7 |
0.873 |
0.023 |
| 13 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.868 |
-0.083 |
| 14 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
0.864 |
-0.018 |
| 15 |
248563_at |
AT5G49690
|
[UDP-Glycosyltransferase superfamily protein] |
0.861 |
-0.013 |
| 16 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.855 |
0.114 |
| 17 |
246149_at |
AT5G19890
|
[Peroxidase superfamily protein] |
0.826 |
-0.072 |
| 18 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.812 |
0.044 |
| 19 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.804 |
0.005 |
| 20 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.803 |
0.129 |
| 21 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
0.801 |
-0.064 |
| 22 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
0.796 |
0.246 |
| 23 |
253301_at |
AT4G33720
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.786 |
-0.017 |
| 24 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
0.784 |
0.366 |
| 25 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.777 |
0.311 |
| 26 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.771 |
0.051 |
| 27 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
0.767 |
-0.002 |
| 28 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.761 |
0.074 |
| 29 |
252045_at |
AT3G52450
|
AtPUB22, PUB22, plant U-box 22 |
0.755 |
0.073 |
| 30 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
0.747 |
0.047 |
| 31 |
260030_at |
AT1G68880
|
AtbZIP, basic leucine-zipper 8, bZIP, basic leucine-zipper 8 |
0.744 |
-0.090 |
| 32 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
0.744 |
0.068 |
| 33 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
0.717 |
0.091 |
| 34 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.691 |
-0.105 |
| 35 |
264375_at |
AT2G25090
|
AtCIPK16, CIPK16, CBL-interacting protein kinase 16, SnRK3.18, SNF1-RELATED PROTEIN KINASE 3.18 |
0.688 |
-0.134 |
| 36 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
0.687 |
-0.017 |
| 37 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
0.687 |
0.015 |
| 38 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.686 |
0.039 |
| 39 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
0.682 |
0.047 |
| 40 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
0.681 |
0.048 |
| 41 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
0.677 |
0.057 |
| 42 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
0.675 |
0.018 |
| 43 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.665 |
0.058 |
| 44 |
263475_at |
AT2G31945
|
unknown |
0.662 |
0.029 |
| 45 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
0.660 |
0.024 |
| 46 |
265853_at |
AT2G42360
|
[RING/U-box superfamily protein] |
0.658 |
0.107 |
| 47 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
0.649 |
0.007 |
| 48 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.642 |
0.033 |
| 49 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.638 |
0.167 |
| 50 |
267035_at |
AT2G38400
|
AGT3, alanine:glyoxylate aminotransferase 3 |
0.638 |
0.088 |
| 51 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
0.637 |
-0.091 |
| 52 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
0.636 |
0.040 |
| 53 |
261606_at |
AT1G49570
|
[Peroxidase superfamily protein] |
0.628 |
-0.053 |
| 54 |
254410_at |
AT4G21410
|
CRK29, cysteine-rich RLK (RECEPTOR-like protein kinase) 29 |
0.627 |
0.038 |
| 55 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
0.625 |
-0.018 |
| 56 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
0.623 |
0.057 |
| 57 |
245076_at |
AT2G23170
|
GH3.3 |
0.619 |
1.412 |
| 58 |
259036_at |
AT3G09220
|
LAC7, laccase 7 |
0.613 |
-0.089 |
| 59 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
0.613 |
-0.176 |
| 60 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.610 |
0.047 |
| 61 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
0.609 |
0.067 |
| 62 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
0.602 |
0.032 |
| 63 |
260744_at |
AT1G15010
|
unknown |
0.597 |
-0.014 |
| 64 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.595 |
-0.080 |
| 65 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
0.591 |
0.057 |
| 66 |
267178_at |
AT2G37750
|
unknown |
0.590 |
-0.039 |
| 67 |
256442_at |
AT3G10930
|
unknown |
0.589 |
0.282 |
| 68 |
259297_at |
AT3G05360
|
AtRLP30, receptor like protein 30, RLP30, receptor like protein 30 |
0.585 |
0.096 |
| 69 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.583 |
-0.116 |
| 70 |
249996_at |
AT5G18600
|
[Thioredoxin superfamily protein] |
0.577 |
0.030 |
| 71 |
262887_at |
AT1G14780
|
[MAC/Perforin domain-containing protein] |
0.569 |
0.005 |
| 72 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
0.564 |
-0.126 |
| 73 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
0.562 |
0.077 |
| 74 |
246984_at |
AT5G67310
|
CYP81G1, cytochrome P450, family 81, subfamily G, polypeptide 1 |
0.552 |
-0.027 |
| 75 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
0.550 |
0.062 |
| 76 |
260522_x_at |
AT2G41730
|
unknown |
0.544 |
-0.074 |
| 77 |
265852_at |
AT2G42350
|
[RING/U-box superfamily protein] |
0.541 |
0.029 |
| 78 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
0.535 |
0.143 |
| 79 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.533 |
-0.061 |
| 80 |
263227_at |
AT1G30750
|
unknown |
0.529 |
-0.063 |
| 81 |
248799_at |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
0.523 |
0.164 |
| 82 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.523 |
0.142 |
| 83 |
267548_at |
AT2G32660
|
AtRLP22, receptor like protein 22, RLP22, receptor like protein 22 |
0.522 |
-0.021 |
| 84 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
0.519 |
0.063 |
| 85 |
248392_at |
AT5G52050
|
[MATE efflux family protein] |
0.519 |
-0.004 |
| 86 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.516 |
-0.064 |
| 87 |
258813_at |
AT3G04060
|
anac046, NAC domain containing protein 46, NAC046, NAC domain containing protein 46 |
0.516 |
0.017 |
| 88 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.513 |
-0.071 |
| 89 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.511 |
0.064 |
| 90 |
261558_at |
AT1G01770
|
unknown |
0.509 |
0.015 |
| 91 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.502 |
-0.087 |
| 92 |
264685_at |
AT1G65610
|
ATGH9A2, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9A2, KOR2, KORRIGAN 2 |
0.502 |
-0.031 |
| 93 |
252530_at |
AT3G46500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.501 |
-0.093 |
| 94 |
255430_at |
AT4G03320
|
AtTic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV, Tic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV |
0.500 |
0.221 |
| 95 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
0.499 |
-0.218 |
| 96 |
250323_at |
AT5G12880
|
[proline-rich family protein] |
0.498 |
0.046 |
| 97 |
248040_at |
AT5G55970
|
[RING/U-box superfamily protein] |
0.498 |
-0.001 |
| 98 |
250182_at |
AT5G14470
|
AtGALK2, GALK2, galactokinase 2 |
0.496 |
0.186 |
| 99 |
266231_at |
AT2G02220
|
ATPSKR1, PHYTOSULFOKIN RECEPTOR 1, PSKR1, phytosulfokin receptor 1 |
0.489 |
0.085 |
| 100 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.488 |
0.032 |
| 101 |
266156_at |
AT2G28110
|
FRA8, FRAGILE FIBER 8, IRX7, IRREGULAR XYLEM 7 |
0.488 |
-0.005 |
| 102 |
256024_at |
AT1G58340
|
BCD1, BUSH-AND-CHLOROTIC-DWARF 1, ZF14, ZRZ, ZRIZI |
0.483 |
-0.065 |
| 103 |
247921_at |
AT5G57660
|
ATCOL5, BBX6, B-box domain protein 6, COL5, CONSTANS-like 5 |
0.481 |
0.019 |
| 104 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
0.478 |
-0.082 |
| 105 |
245353_at |
AT4G16000
|
unknown |
0.478 |
0.037 |
| 106 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.476 |
0.048 |
| 107 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
0.476 |
-0.034 |
| 108 |
254667_at |
AT4G18280
|
[glycine-rich cell wall protein-related] |
0.474 |
-0.021 |
| 109 |
258110_at |
AT3G14610
|
CYP72A7, cytochrome P450, family 72, subfamily A, polypeptide 7 |
0.473 |
0.081 |
| 110 |
253125_at |
AT4G36040
|
DJC23, DNA J protein C23, J11, DnaJ11 |
0.472 |
0.027 |
| 111 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.469 |
0.018 |
| 112 |
259481_at |
AT1G18970
|
GLP4, germin-like protein 4 |
0.469 |
0.033 |
| 113 |
261544_at |
AT1G63540
|
[hydroxyproline-rich glycoprotein family protein] |
0.469 |
-0.072 |
| 114 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-1.107 |
-0.198 |
| 115 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.963 |
-0.045 |
| 116 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
-0.939 |
-0.284 |
| 117 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
-0.938 |
-0.186 |
| 118 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.904 |
-0.024 |
| 119 |
266353_at |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
-0.902 |
-0.074 |
| 120 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-0.864 |
-0.036 |
| 121 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.844 |
-0.118 |
| 122 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-0.811 |
-0.088 |
| 123 |
261660_at |
AT1G18370
|
ATNACK1, ARABIDOPSIS NPK1-ACTIVATING KINESIN 1, HIK, HINKEL, NACK1, NPK1-ACTIVATING KINESIN 1 |
-0.758 |
-0.130 |
| 124 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
-0.749 |
-0.095 |
| 125 |
264271_at |
AT1G60270
|
BGLU6, beta glucosidase 6 |
-0.745 |
-0.094 |
| 126 |
251226_at |
AT3G62680
|
ATPRP3, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 3, PRP3, proline-rich protein 3 |
-0.731 |
0.039 |
| 127 |
262651_at |
AT1G14100
|
FUT8, fucosyltransferase 8 |
-0.725 |
-0.028 |
| 128 |
254233_at |
AT4G23800
|
3xHMG-box2, 3xHigh Mobility Group-box2 |
-0.710 |
-0.044 |
| 129 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.695 |
-0.142 |
| 130 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
-0.659 |
-0.048 |
| 131 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
-0.653 |
-0.220 |
| 132 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.651 |
-0.057 |
| 133 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.643 |
-0.213 |
| 134 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
-0.642 |
-0.096 |
| 135 |
265305_at |
AT2G20340
|
AAS, aromatic aldehyde synthase, AtAAS |
-0.636 |
-0.085 |
| 136 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.635 |
-0.169 |
| 137 |
264802_at |
AT1G08560
|
ATSYP111, KN, KNOLLE, SYP111, syntaxin of plants 111 |
-0.635 |
-0.133 |
| 138 |
247977_at |
AT5G56850
|
unknown |
-0.630 |
0.023 |
| 139 |
261562_at |
AT1G01750
|
ADF11, actin depolymerizing factor 11 |
-0.627 |
-0.029 |
| 140 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.622 |
-0.182 |
| 141 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
-0.620 |
-0.203 |
| 142 |
249545_at |
AT5G38030
|
[MATE efflux family protein] |
-0.606 |
-0.044 |
| 143 |
248057_at |
AT5G55520
|
unknown |
-0.604 |
-0.114 |
| 144 |
246415_at |
AT5G17160
|
unknown |
-0.603 |
-0.128 |
| 145 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.597 |
-0.140 |
| 146 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.595 |
-0.167 |
| 147 |
263902_at |
AT2G36230
|
APG10, ALBINO AND PALE GREEN 10, HISN3 |
-0.587 |
0.032 |
| 148 |
248624_at |
AT5G48790
|
unknown |
-0.585 |
0.040 |
| 149 |
256675_at |
AT3G52170
|
[DNA binding] |
-0.580 |
0.017 |
| 150 |
256244_at |
AT3G12520
|
SULTR4;2, sulfate transporter 4;2 |
-0.571 |
-0.088 |
| 151 |
253337_at |
AT4G33470
|
ATHDA14, HDA14, histone deacetylase 14 |
-0.570 |
0.099 |
| 152 |
253947_at |
AT4G26760
|
MAP65-2, microtubule-associated protein 65-2 |
-0.565 |
-0.071 |
| 153 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
-0.564 |
-0.116 |
| 154 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.564 |
0.013 |
| 155 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
-0.559 |
-0.121 |
| 156 |
265957_at |
AT2G37300
|
ABCI16, ATP-binding cassette I16 |
-0.547 |
-0.057 |
| 157 |
255265_at |
AT4G05190
|
ATK5, kinesin 5 |
-0.542 |
-0.076 |
| 158 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.541 |
-0.025 |
| 159 |
253244_at |
AT4G34580
|
AtSFH1, COW1, CAN OF WORMS1, SRH1, SHORT ROOT HAIR 1 |
-0.533 |
-0.030 |
| 160 |
263597_at |
AT2G01870
|
unknown |
-0.530 |
0.057 |
| 161 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.520 |
-0.153 |
| 162 |
256864_at |
AT3G23890
|
ATTOPII, TOPII, topoisomerase II |
-0.507 |
-0.056 |
| 163 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
-0.503 |
-0.041 |
| 164 |
265083_at |
AT1G03820
|
unknown |
-0.497 |
-0.089 |
| 165 |
257474_at |
AT1G80850
|
[DNA glycosylase superfamily protein] |
-0.496 |
0.049 |
| 166 |
266330_at |
AT2G01530
|
MLP329, MLP-like protein 329, ZCE2, (Zusammen-CA)-enhanced 2 |
-0.496 |
-0.021 |
| 167 |
258471_at |
AT3G06030
|
ANP3, NPK1-related protein kinase 3, AtANP3, MAPKKK12, NP3, NPK1-related protein kinase 3 |
-0.484 |
-0.093 |
| 168 |
257714_at |
AT3G27360
|
H3.1, histone 3.1 |
-0.469 |
-0.009 |
| 169 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
-0.468 |
-0.123 |
| 170 |
258264_at |
AT3G15790
|
ATMBD11, MBD11, methyl-CPG-binding domain 11 |
-0.460 |
-0.069 |
| 171 |
250689_at |
AT5G06610
|
unknown |
-0.459 |
0.000 |
| 172 |
261593_at |
AT1G33170
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.458 |
-0.148 |
| 173 |
246487_at |
AT5G16030
|
unknown |
-0.446 |
-0.087 |
| 174 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
-0.442 |
-0.066 |
| 175 |
255664_at |
AT4G00440
|
TRM15, TON1 Recruiting Motif 15 |
-0.442 |
-0.035 |
| 176 |
257663_at |
AT3G20260
|
[FUNCTIONS IN: structural constituent of ribosome] |
-0.439 |
-0.049 |
| 177 |
247331_at |
AT5G63530
|
ATFP3, ARABIDOPSIS THALIANA FARNESYLATED PROTEIN 3, FP3, farnesylated protein 3 |
-0.438 |
-0.012 |
| 178 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
-0.438 |
-0.044 |
| 179 |
257875_at |
AT3G17120
|
unknown |
-0.433 |
-0.046 |
| 180 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.428 |
-0.018 |
| 181 |
263441_at |
AT2G28620
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.427 |
-0.019 |
| 182 |
264839_at |
AT1G03630
|
POR C, protochlorophyllide oxidoreductase C, PORC |
-0.427 |
0.035 |
| 183 |
246557_at |
AT5G15510
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.426 |
-0.103 |
| 184 |
245816_at |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
-0.421 |
0.030 |
| 185 |
262750_at |
AT1G28710
|
[Nucleotide-diphospho-sugar transferase family protein] |
-0.421 |
0.038 |
| 186 |
259851_at |
AT1G72250
|
[Di-glucose binding protein with Kinesin motor domain] |
-0.420 |
-0.139 |
| 187 |
245576_at |
AT4G14770
|
ATTCX2, TESMIN/TSO1-LIKE CXC 2, TCX2, TESMIN/TSO1-like CXC 2 |
-0.419 |
0.107 |
| 188 |
256052_at |
AT1G06960
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.411 |
0.126 |
| 189 |
247486_at |
AT5G62140
|
unknown |
-0.411 |
-0.138 |
| 190 |
255236_at |
AT4G05520
|
ATEHD2, EPS15 homology domain 2, EHD2, EPS15 homology domain 2 |
-0.408 |
-0.048 |
| 191 |
261806_at |
AT1G30510
|
ATRFNR2, root FNR 2, RFNR2, root FNR 2 |
-0.407 |
0.181 |
| 192 |
261536_at |
AT1G01790
|
ATKEA1, K+ EFFLUX ANTIPORTER 1, KEA1, K+ efflux antiporter 1 |
-0.403 |
0.008 |
| 193 |
254485_at |
AT4G20760
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.401 |
0.004 |
| 194 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
-0.400 |
-0.168 |
| 195 |
249644_at |
AT5G37010
|
unknown |
-0.399 |
-0.070 |
| 196 |
266372_at |
AT2G41310
|
ARR8, RESPONSE REGULATOR 8, ATRR3, response regulator 3, RR3, response regulator 3 |
-0.396 |
-0.160 |
| 197 |
257115_at |
AT3G20150
|
[Kinesin motor family protein] |
-0.393 |
-0.009 |
| 198 |
260801_at |
AT1G78430
|
RIP4, ROP interactive partner 4 |
-0.393 |
-0.098 |
| 199 |
257848_at |
AT3G13030
|
[hAT transposon superfamily protein] |
-0.392 |
-0.007 |
| 200 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.392 |
-0.132 |
| 201 |
254928_at |
AT4G11410
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.388 |
0.022 |
| 202 |
263607_at |
AT2G16270
|
unknown |
-0.384 |
-0.044 |
| 203 |
254791_at |
AT4G12910
|
scpl20, serine carboxypeptidase-like 20 |
-0.383 |
-0.075 |
| 204 |
255597_at |
AT4G01730
|
[DHHC-type zinc finger family protein] |
-0.383 |
-0.062 |
| 205 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.380 |
0.001 |
| 206 |
249046_at |
AT5G44400
|
[FAD-binding Berberine family protein] |
-0.379 |
-0.118 |
| 207 |
246899_at |
AT5G25590
|
[INVOLVED IN: N-terminal protein myristoylation] |
-0.378 |
-0.071 |
| 208 |
260034_at |
AT1G68810
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.376 |
0.038 |
| 209 |
263677_at |
AT1G04520
|
PDLP2, plasmodesmata-located protein 2 |
-0.376 |
-0.000 |
| 210 |
245712_at |
AT5G04360
|
ATLDA, limit dextrinase, ATPU1, PULLULANASE 1, LDA, limit dextrinase, PU1, PULLULANASE 1 |
-0.376 |
0.087 |
| 211 |
262081_at |
AT1G59540
|
ZCF125 |
-0.371 |
-0.123 |
| 212 |
254505_at |
AT4G19985
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.369 |
-0.009 |
| 213 |
262693_at |
AT1G62780
|
unknown |
-0.368 |
-0.003 |
| 214 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
-0.365 |
-0.297 |
| 215 |
247716_at |
AT5G59350
|
unknown |
-0.359 |
-0.111 |
| 216 |
258013_at |
AT3G19320
|
[Leucine-rich repeat (LRR) family protein] |
-0.358 |
-0.051 |
| 217 |
267402_at |
AT2G26180
|
IQD6, IQ-domain 6 |
-0.358 |
0.041 |
| 218 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.354 |
-0.130 |
| 219 |
264360_at |
AT1G03310
|
AtBE2, ATISA2, ARABIDOPSIS THALIANA ISOAMYLASE 2, BE2, BRANCHING ENZYME 2, DBE1, debranching enzyme 1, ISA2 |
-0.353 |
0.075 |
| 220 |
262773_at |
AT1G13220
|
CRWN2, CROWDED NUCLEI 2, LINC2, LITTLE NUCLEI2 |
-0.352 |
0.009 |
| 221 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.351 |
-0.281 |
| 222 |
248658_at |
AT5G48600
|
ATCAP-C, ARABIDOPSIS THALIANA CHROMOSOME ASSOCIATED PROTEIN-C, ATSMC3, structural maintenance of chromosome 3, ATSMC4, ARABIDOPSIS THALIANA STRUCTURAL MAINTENANCE OF CHROMOSOME 4, SMC3, structural maintenance of chromosome 3 |
-0.349 |
-0.067 |
| 223 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
-0.349 |
-0.035 |
| 224 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
-0.348 |
-0.004 |
| 225 |
253978_at |
AT4G26660
|
unknown |
-0.345 |
-0.104 |