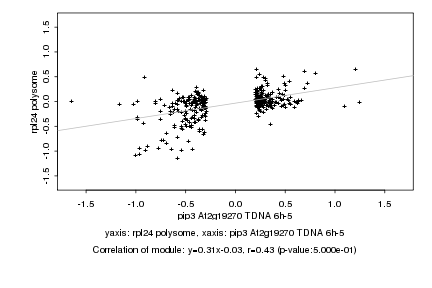
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
pip3 At2g19270 TDNA 6h-5 |
Log2 signal ratio
rpl24 polysome |
| 1 |
246671_at |
AT5G30450
|
[CACTA-like transposase family (Ptta/En/Spm), has a 4.3e-19 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
1.237 |
-0.007 |
| 2 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
1.198 |
0.665 |
| 3 |
255471_at |
AT4G03050
|
AOP3 |
1.093 |
-0.092 |
| 4 |
250135_at |
AT5G15360
|
unknown |
0.804 |
0.578 |
| 5 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
0.718 |
0.370 |
| 6 |
256878_at |
AT3G26460
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.691 |
0.276 |
| 7 |
253532_at |
AT4G31570
|
unknown |
0.690 |
0.613 |
| 8 |
260723_at |
AT1G48070
|
[Thioredoxin superfamily protein] |
0.656 |
0.030 |
| 9 |
267330_at |
AT2G19270
|
unknown |
0.632 |
0.026 |
| 10 |
248810_at |
AT5G47280
|
ADR1-L3, ADR1-like 3 |
0.624 |
0.019 |
| 11 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
0.620 |
-0.037 |
| 12 |
249461_at |
AT5G39640
|
[Putative endonuclease or glycosyl hydrolase] |
0.605 |
-0.006 |
| 13 |
249053_at |
AT5G44440
|
[FAD-binding Berberine family protein] |
0.591 |
0.020 |
| 14 |
257417_at |
AT1G10110
|
[F-box family protein] |
0.585 |
-0.118 |
| 15 |
249970_at |
AT5G19100
|
[Eukaryotic aspartyl protease family protein] |
0.551 |
0.086 |
| 16 |
256859_at |
AT3G22940
|
[F-box associated ubiquitination effector family protein] |
0.547 |
-0.044 |
| 17 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.544 |
-0.057 |
| 18 |
254350_at |
AT4G22280
|
[F-box/RNI-like superfamily protein] |
0.535 |
0.408 |
| 19 |
257859_at |
AT3G12955
|
SAUR74, SMALL AUXIN UPREGULATED RNA 74 |
0.535 |
-0.027 |
| 20 |
248407_at |
AT5G51500
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.531 |
0.037 |
| 21 |
258670_at |
AT3G08810
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.516 |
0.048 |
| 22 |
246537_at |
AT5G15940
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.500 |
-0.119 |
| 23 |
246814_at |
AT5G27200
|
ACP5, acyl carrier protein 5 |
0.494 |
0.327 |
| 24 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.494 |
0.231 |
| 25 |
259481_at |
AT1G18970
|
GLP4, germin-like protein 4 |
0.492 |
-0.056 |
| 26 |
264016_at |
AT2G21220
|
SAUR12, SMALL AUXIN UPREGULATED RNA 12 |
0.490 |
0.138 |
| 27 |
265483_at |
AT2G15790
|
CYP40, CYCLOPHILIN 40, SQN, SQUINT |
0.484 |
0.378 |
| 28 |
256736_at |
AT3G29410
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.480 |
0.044 |
| 29 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
0.474 |
0.511 |
| 30 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.473 |
-0.075 |
| 31 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.463 |
0.145 |
| 32 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
0.462 |
0.021 |
| 33 |
265587_at |
AT2G19980
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.454 |
-0.088 |
| 34 |
247047_at |
AT5G66650
|
unknown |
0.452 |
0.080 |
| 35 |
255111_at |
AT4G08780
|
[Peroxidase superfamily protein] |
0.441 |
0.170 |
| 36 |
253945_at |
AT4G27050
|
[F-box/RNI-like superfamily protein] |
0.432 |
-0.020 |
| 37 |
250576_at |
AT5G08250
|
[Cytochrome P450 superfamily protein] |
0.429 |
0.092 |
| 38 |
260475_at |
AT1G11080
|
scpl31, serine carboxypeptidase-like 31 |
0.428 |
0.262 |
| 39 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.412 |
-0.005 |
| 40 |
254907_at |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.404 |
0.094 |
| 41 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
0.393 |
-0.014 |
| 42 |
264019_at |
AT2G21130
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.385 |
-0.072 |
| 43 |
263672_at |
AT2G04820
|
[copia-like retrotransposon family, has a 2.8e-214 P-value blast match to GB:AAC02666 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)] |
0.372 |
0.003 |
| 44 |
251824_at |
AT3G55090
|
ABCG16, ATP-binding cassette G16 |
0.371 |
-0.124 |
| 45 |
249321_at |
AT5G40920
|
[pseudogene] |
0.364 |
0.045 |
| 46 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.364 |
0.186 |
| 47 |
261385_at |
AT1G05450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.358 |
-0.105 |
| 48 |
259015_at |
AT3G07350
|
unknown |
0.354 |
-0.032 |
| 49 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
0.349 |
-0.063 |
| 50 |
260146_at |
AT1G52770
|
[Phototropic-responsive NPH3 family protein] |
0.349 |
0.156 |
| 51 |
246256_at |
AT4G36770
|
[UDP-Glycosyltransferase superfamily protein] |
0.349 |
0.017 |
| 52 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
0.347 |
-0.004 |
| 53 |
254208_at |
AT4G24175
|
unknown |
0.345 |
0.080 |
| 54 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.343 |
-0.448 |
| 55 |
254954_at |
AT4G10910
|
unknown |
0.343 |
0.160 |
| 56 |
254634_at |
AT4G18650
|
[transcription factor-related] |
0.341 |
0.073 |
| 57 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.340 |
-0.029 |
| 58 |
254193_at |
AT4G23870
|
unknown |
0.337 |
-0.017 |
| 59 |
259364_at |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
0.334 |
0.029 |
| 60 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
0.331 |
-0.000 |
| 61 |
252488_at |
AT3G46700
|
[UDP-Glycosyltransferase superfamily protein] |
0.331 |
-0.089 |
| 62 |
250127_at |
AT5G16380
|
unknown |
0.330 |
0.155 |
| 63 |
246831_at |
AT5G26340
|
ATSTP13, SUGAR TRANSPORT PROTEIN 13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
0.329 |
0.154 |
| 64 |
263198_at |
AT1G53990
|
GLIP3, GDSL-motif lipase 3 |
0.328 |
-0.019 |
| 65 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.327 |
-0.005 |
| 66 |
263953_at |
AT2G36050
|
ATOFP15, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 15, OFP15, ovate family protein 15 |
0.326 |
-0.011 |
| 67 |
256138_at |
AT1G48670
|
[auxin-responsive GH3 family protein] |
0.320 |
-0.063 |
| 68 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
0.317 |
0.339 |
| 69 |
252533_at |
AT3G46110
|
[LOCATED IN: plasma membrane] |
0.316 |
0.113 |
| 70 |
249843_at |
AT5G23570
|
ATSGS3, SUPPRESSOR OF GENE SILENCING 3, SGS3, SUPPRESSOR OF GENE SILENCING 3 |
0.313 |
0.369 |
| 71 |
267429_at |
AT2G34850
|
MEE25, maternal effect embryo arrest 25 |
0.313 |
0.129 |
| 72 |
256097_at |
AT1G13670
|
unknown |
0.312 |
0.101 |
| 73 |
264167_at |
AT1G02060
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.311 |
-0.062 |
| 74 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
0.309 |
-0.078 |
| 75 |
250597_at |
AT5G07680
|
ANAC079, Arabidopsis NAC domain containing protein 79, ANAC080, NAC domain containing protein 80, ATNAC4, NAC080, NAC domain containing protein 80, NAC4 |
0.309 |
0.062 |
| 76 |
254572_at |
AT4G19380
|
[Long-chain fatty alcohol dehydrogenase family protein] |
0.305 |
-0.149 |
| 77 |
248619_at |
AT5G49630
|
AAP6, amino acid permease 6 |
0.298 |
0.018 |
| 78 |
261001_at |
AT1G26530
|
[PIN domain-like family protein] |
0.297 |
0.431 |
| 79 |
247833_at |
AT5G58575
|
unknown |
0.296 |
0.132 |
| 80 |
256489_at |
AT1G31550
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.292 |
0.474 |
| 81 |
255751_at |
AT1G31950
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.291 |
0.046 |
| 82 |
252694_at |
AT3G43630
|
[Vacuolar iron transporter (VIT) family protein] |
0.287 |
-0.030 |
| 83 |
266259_at |
AT2G27830
|
unknown |
0.286 |
0.057 |
| 84 |
249489_at |
AT5G39090
|
[HXXXD-type acyl-transferase family protein] |
0.285 |
0.035 |
| 85 |
264375_at |
AT2G25090
|
AtCIPK16, CIPK16, CBL-interacting protein kinase 16, SnRK3.18, SNF1-RELATED PROTEIN KINASE 3.18 |
0.280 |
0.240 |
| 86 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
0.278 |
-0.219 |
| 87 |
265387_at |
AT2G20670
|
unknown |
0.275 |
0.029 |
| 88 |
260237_at |
AT1G74430
|
ATMYB95, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 95, ATMYBCP66, ARABIDOPSIS THALIANA MYB DOMAIN CONTAINING PROTEIN 66, MYB95, myb domain protein 95 |
0.275 |
0.036 |
| 89 |
265732_at |
AT2G01300
|
unknown |
0.275 |
0.002 |
| 90 |
247317_at |
AT5G64060
|
anac103, NAC domain containing protein 103, NAC103, NAC domain containing protein 103 |
0.273 |
0.499 |
| 91 |
266640_at |
AT2G35585
|
unknown |
0.273 |
-0.109 |
| 92 |
266900_at |
AT2G34610
|
unknown |
0.271 |
0.012 |
| 93 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.271 |
-0.005 |
| 94 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
0.270 |
0.072 |
| 95 |
264716_at |
AT1G70170
|
MMP, matrix metalloproteinase |
0.269 |
0.090 |
| 96 |
251249_at |
AT3G62160
|
[HXXXD-type acyl-transferase family protein] |
0.269 |
-0.070 |
| 97 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.267 |
0.019 |
| 98 |
262665_at |
AT1G14070
|
FUT7, fucosyltransferase 7 |
0.263 |
0.320 |
| 99 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.262 |
0.180 |
| 100 |
265877_at |
AT2G42380
|
ATBZIP34, BZIP34 |
0.261 |
-0.053 |
| 101 |
255742_at |
AT1G25560
|
AtTEM1, EDF1, ETHYLENE RESPONSE DNA BINDING FACTOR 1, TEM1, TEMPRANILLO 1 |
0.257 |
0.142 |
| 102 |
246817_at |
AT5G27240
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.255 |
0.227 |
| 103 |
252557_at |
AT3G45960
|
ATEXLA3, expansin-like A3, ATEXPL3, ATHEXP BETA 2.3, EXLA3, expansin-like A3, EXPL3 |
0.254 |
0.032 |
| 104 |
267300_at |
AT2G30140
|
UGT87A2, UDP-glucosyl transferase 87A2 |
0.253 |
0.199 |
| 105 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
0.252 |
-0.055 |
| 106 |
262529_at |
AT1G17250
|
AtRLP3, receptor like protein 3, RFO2, RESISTANCE TO FUSARIUM OXYSPORUM 2, RLP3, receptor like protein 3 |
0.252 |
0.313 |
| 107 |
262126_at |
AT1G59620
|
CW9 |
0.251 |
-0.095 |
| 108 |
265290_at |
AT2G22590
|
[UDP-Glycosyltransferase superfamily protein] |
0.251 |
-0.017 |
| 109 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.249 |
0.005 |
| 110 |
250589_at |
AT5G07700
|
AtMYB76, myb domain protein 76, MYB76, myb domain protein 76 |
0.249 |
0.112 |
| 111 |
261265_at |
AT1G26800
|
[RING/U-box superfamily protein] |
0.248 |
-0.041 |
| 112 |
264752_at |
AT1G23010
|
LPR1, Low Phosphate Root1 |
0.248 |
-0.183 |
| 113 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
0.247 |
0.055 |
| 114 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
0.247 |
-0.041 |
| 115 |
260169_at |
AT1G71990
|
ATFT4, ARABIDOPSIS FUCOSYLTRANSFERASE 4, ATFUT13, FT4-M, FUCTC, FUT13, fucosyltransferase 13 |
0.246 |
0.127 |
| 116 |
249881_at |
AT5G23190
|
CYP86B1, cytochrome P450, family 86, subfamily B, polypeptide 1 |
0.243 |
-0.092 |
| 117 |
265276_at |
AT2G28400
|
unknown |
0.243 |
0.272 |
| 118 |
246996_at |
AT5G67420
|
ASL39, ASYMMETRIC LEAVES2-LIKE 39, LBD37, LOB domain-containing protein 37 |
0.242 |
-0.066 |
| 119 |
252707_at |
AT3G43790
|
ZIFL2, zinc induced facilitator-like 2 |
0.242 |
0.043 |
| 120 |
255280_at |
AT4G04960
|
LecRK-VII.1, L-type lectin receptor kinase VII.1 |
0.241 |
-0.033 |
| 121 |
254919_at |
AT4G11360
|
RHA1B, RING-H2 finger A1B |
0.241 |
0.060 |
| 122 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
0.240 |
-0.035 |
| 123 |
252395_at |
AT3G47950
|
AHA4, H(+)-ATPase 4, HA4, H(+)-ATPase 4 |
0.240 |
-0.003 |
| 124 |
263005_at |
AT1G54540
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.239 |
0.034 |
| 125 |
253317_at |
AT4G33960
|
unknown |
0.239 |
0.091 |
| 126 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
0.235 |
0.563 |
| 127 |
254037_at |
AT4G25760
|
ATGDU2, glutamine dumper 2, GDU2, glutamine dumper 2 |
0.234 |
-0.168 |
| 128 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
0.234 |
-0.055 |
| 129 |
251280_at |
AT3G61810
|
[Glycosyl hydrolase family 17 protein] |
0.234 |
-0.066 |
| 130 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.233 |
-0.153 |
| 131 |
259979_at |
AT1G76600
|
unknown |
0.233 |
0.003 |
| 132 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.233 |
0.265 |
| 133 |
260415_at |
AT1G69790
|
[Protein kinase superfamily protein] |
0.232 |
-0.017 |
| 134 |
246251_at |
AT4G37220
|
[Cold acclimation protein WCOR413 family] |
0.231 |
0.091 |
| 135 |
266108_at |
AT2G37900
|
[Major facilitator superfamily protein] |
0.231 |
0.018 |
| 136 |
252193_at |
AT3G50060
|
MYB77, myb domain protein 77 |
0.230 |
0.016 |
| 137 |
259478_at |
AT1G18980
|
[RmlC-like cupins superfamily protein] |
0.229 |
-0.029 |
| 138 |
252483_at |
AT3G46600
|
[GRAS family transcription factor] |
0.228 |
-0.102 |
| 139 |
266274_at |
AT2G29380
|
HAI3, highly ABA-induced PP2C gene 3 |
0.228 |
0.003 |
| 140 |
259548_at |
AT1G35260
|
MLP165, MLP-like protein 165 |
0.228 |
0.055 |
| 141 |
249274_at |
AT5G41860
|
unknown |
0.227 |
-0.289 |
| 142 |
253152_at |
AT4G35690
|
unknown |
0.224 |
-0.021 |
| 143 |
257274_at |
AT3G14510
|
[Polyprenyl synthetase family protein] |
0.224 |
-0.019 |
| 144 |
245252_at |
AT4G17500
|
ATERF-1, ethylene responsive element binding factor 1, ERF-1, ethylene responsive element binding factor 1 |
0.223 |
0.104 |
| 145 |
262656_at |
AT1G14200
|
[RING/U-box superfamily protein] |
0.220 |
0.106 |
| 146 |
262233_at |
AT1G48310
|
CHA18, CHR18, chromatin remodeling factor18 |
0.219 |
0.042 |
| 147 |
260522_x_at |
AT2G41730
|
unknown |
0.218 |
0.106 |
| 148 |
266344_at |
AT2G01580
|
unknown |
0.218 |
-0.069 |
| 149 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
0.217 |
0.026 |
| 150 |
245912_at |
AT5G19600
|
SULTR3;5, sulfate transporter 3;5 |
0.216 |
0.060 |
| 151 |
247683_at |
AT5G59660
|
[Leucine-rich repeat protein kinase family protein] |
0.216 |
0.086 |
| 152 |
256098_at |
AT1G13700
|
PGL1, 6-phosphogluconolactonase 1 |
0.215 |
0.214 |
| 153 |
260803_at |
AT1G78340
|
ATGSTU22, glutathione S-transferase TAU 22, GSTU22, glutathione S-transferase TAU 22 |
0.215 |
0.043 |
| 154 |
245078_at |
AT2G23340
|
DEAR3, DREB and EAR motif protein 3 |
0.215 |
0.027 |
| 155 |
255403_at |
AT4G03400
|
DFL2, DWARF IN LIGHT 2, GH3-10 |
0.214 |
0.267 |
| 156 |
249771_at |
AT5G24080
|
[Protein kinase superfamily protein] |
0.214 |
-0.103 |
| 157 |
257679_at |
AT3G20470
|
ATGRP-5, ATGRP5, GRP-5, GLYCINE-RICH PROTEIN 5, GRP5, glycine-rich protein 5 |
0.212 |
0.229 |
| 158 |
262737_at |
AT1G28560
|
SRD2, SHOOT REDIFFERENTIATION DEFECTIVE 2 |
0.212 |
0.178 |
| 159 |
260037_at |
AT1G68840
|
AtRAV2, EDF2, ETHYLENE RESPONSE DNA BINDING FACTOR 2, RAP2.8, RELATED TO AP2 8, RAV2, related to ABI3/VP1 2, TEM2, TEMPRANILLO 2 |
0.212 |
-0.117 |
| 160 |
249601_at |
AT5G37980
|
[Zinc-binding dehydrogenase family protein] |
0.211 |
0.067 |
| 161 |
260264_at |
AT1G68500
|
unknown |
0.209 |
-0.087 |
| 162 |
247827_at |
AT5G58500
|
LSH5, LIGHT SENSITIVE HYPOCOTYLS 5 |
0.209 |
0.045 |
| 163 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.208 |
-0.032 |
| 164 |
267381_at |
AT2G26190
|
[calmodulin-binding family protein] |
0.208 |
-0.063 |
| 165 |
251458_at |
AT3G60170
|
[copia-like retrotransposon family, has a 7.6e-229 P-value blast match to gb|AAO73527.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
0.206 |
0.003 |
| 166 |
246253_at |
AT4G37260
|
ATMYB73, MYB73, myb domain protein 73 |
0.206 |
0.009 |
| 167 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
0.206 |
0.497 |
| 168 |
256852_at |
AT3G18610
|
ATNUC-L2, nucleolin like 2, NUC-L2, nucleolin like 2, PARLL1, PARALLEL1-LIKE 1 |
0.205 |
0.655 |
| 169 |
245581_at |
AT4G14840
|
unknown |
0.205 |
0.188 |
| 170 |
247015_at |
AT5G66960
|
[Prolyl oligopeptidase family protein] |
0.205 |
0.153 |
| 171 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
0.202 |
-0.234 |
| 172 |
250165_at |
AT5G15290
|
CASP5, Casparian strip membrane domain protein 5 |
0.202 |
-0.014 |
| 173 |
258939_at |
AT3G10020
|
unknown |
0.202 |
0.119 |
| 174 |
257694_at |
AT3G12860
|
[NOP56-like pre RNA processing ribonucleoprotein] |
0.201 |
0.251 |
| 175 |
246700_at |
AT5G28030
|
DES1, L-cysteine desulfhydrase 1 |
0.201 |
0.151 |
| 176 |
253839_at |
AT4G27890
|
[HSP20-like chaperones superfamily protein] |
0.201 |
0.051 |
| 177 |
247617_at |
AT5G60270
|
LecRK-I.7, L-type lectin receptor kinase I.7 |
0.200 |
0.188 |
| 178 |
255257_at |
AT4G05050
|
UBQ11, ubiquitin 11 |
-1.654 |
-0.008 |
| 179 |
254250_at |
AT4G23290
|
CRK21, cysteine-rich RLK (RECEPTOR-like protein kinase) 21 |
-1.172 |
-0.045 |
| 180 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
-1.033 |
-0.057 |
| 181 |
263023_at |
AT1G23960
|
unknown |
-1.004 |
-1.087 |
| 182 |
254251_at |
AT4G23300
|
CRK22, cysteine-rich RLK (RECEPTOR-like protein kinase) 22 |
-0.993 |
-0.361 |
| 183 |
256096_at |
AT1G13650
|
unknown |
-0.989 |
0.002 |
| 184 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-0.986 |
-0.310 |
| 185 |
256497_at |
AT1G31580
|
CXC750, ECS1 |
-0.972 |
-1.054 |
| 186 |
252703_at |
AT3G43740
|
[Leucine-rich repeat (LRR) family protein] |
-0.966 |
-0.930 |
| 187 |
259573_at |
AT1G20390
|
[gypsy-like retrotransposon family, has a 1.5e-251 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
-0.931 |
-0.433 |
| 188 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
-0.923 |
0.501 |
| 189 |
252648_at |
AT3G44630
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.908 |
-0.987 |
| 190 |
254585_at |
AT4G19500
|
[nucleoside-triphosphatases] |
-0.889 |
-0.896 |
| 191 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
-0.806 |
-0.020 |
| 192 |
260541_at |
AT2G43530
|
[Scorpion toxin-like knottin superfamily protein] |
-0.803 |
0.015 |
| 193 |
248415_at |
AT5G51620
|
[Uncharacterised protein family (UPF0172)] |
-0.779 |
-0.933 |
| 194 |
260004_at |
AT1G67860
|
unknown |
-0.761 |
-0.196 |
| 195 |
267349_at |
AT2G40010
|
[Ribosomal protein L10 family protein] |
-0.757 |
-0.358 |
| 196 |
250474_at |
AT5G10230
|
ANN7, ANNEXIN 7, ANNAT7, annexin 7 |
-0.756 |
0.052 |
| 197 |
260312_at |
AT1G63880
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.752 |
-0.774 |
| 198 |
250063_at |
AT5G17880
|
CSA1, constitutive shade-avoidance1 |
-0.725 |
-0.782 |
| 199 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.716 |
-0.078 |
| 200 |
250064_at |
AT5G17890
|
CHS3, CHILLING SENSITIVE 3, DAR4, DA1-related protein 4 |
-0.701 |
-0.829 |
| 201 |
245451_at |
AT4G16890
|
BAL, BALL, SNC1, SUPPRESSOR OF NPR1-1, CONSTITUTIVE 1 |
-0.700 |
-0.628 |
| 202 |
258424_at |
AT3G16750
|
unknown |
-0.667 |
-0.103 |
| 203 |
264101_at |
AT1G79000
|
ATHAC1, ARABIDOPSIS HISTONE ACETYLTRANSFERASE OF THE CBP FAMILY 1, ATHPCAT2, ARABIDOPSIS THALIANA P300/CBP ACETYLTRANSFERASE-RELATED PROTEIN 2, HAC1, histone acetyltransferase of the CBP family 1, PCAT2, P300/CBP ACETYLTRANSFERASE-RELATED PROTEIN 2 |
-0.656 |
-0.249 |
| 204 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
-0.653 |
-0.194 |
| 205 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
-0.643 |
-0.955 |
| 206 |
245456_at |
AT4G16950
|
RPP5, RECOGNITION OF PERONOSPORA PARASITICA 5 |
-0.642 |
-0.034 |
| 207 |
248978_at |
AT5G45070
|
AtPP2-A8, phloem protein 2-A8, PP2-A8, phloem protein 2-A8 |
-0.641 |
-0.085 |
| 208 |
262910_at |
AT1G59710
|
unknown |
-0.640 |
0.231 |
| 209 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
-0.626 |
-0.220 |
| 210 |
262908_at |
AT1G59900
|
AT-E1 ALPHA, pyruvate dehydrogenase complex E1 alpha subunit, E1 ALPHA, pyruvate dehydrogenase complex E1 alpha subunit |
-0.622 |
-0.149 |
| 211 |
264751_at |
AT1G23020
|
ATFRO3, FERRIC REDUCTION OXIDASE 3, FRO3, ferric reduction oxidase 3 |
-0.621 |
-0.476 |
| 212 |
246888_at |
AT5G26270
|
unknown |
-0.614 |
-0.518 |
| 213 |
254276_at |
AT4G22820
|
[A20/AN1-like zinc finger family protein] |
-0.606 |
-0.022 |
| 214 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.592 |
0.024 |
| 215 |
246375_at |
AT1G51830
|
[Leucine-rich repeat protein kinase family protein] |
-0.588 |
-0.046 |
| 216 |
252485_at |
AT3G46530
|
RPP13, RECOGNITION OF PERONOSPORA PARASITICA 13 |
-0.588 |
-0.717 |
| 217 |
245448_at |
AT4G16860
|
RPP4, recognition of peronospora parasitica 4 |
-0.587 |
-1.132 |
| 218 |
258059_at |
AT3G29035
|
ANAC059, Arabidopsis NAC domain containing protein 59, ATNAC3, NAC domain containing protein 3, NAC3, NAC domain containing protein 3, ORS1, ORE1 SISTER1 |
-0.585 |
0.179 |
| 219 |
249320_at |
AT5G40910
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.585 |
-0.158 |
| 220 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
-0.576 |
-0.183 |
| 221 |
249934_at |
AT5G22410
|
RHS18, root hair specific 18 |
-0.572 |
0.027 |
| 222 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
-0.565 |
0.062 |
| 223 |
262010_at |
AT1G35612
|
[expressed protein] |
-0.550 |
-0.978 |
| 224 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.549 |
0.016 |
| 225 |
246478_at |
AT5G15980
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.549 |
-0.218 |
| 226 |
257272_at |
AT3G28130
|
UMAMIT44, Usually multiple acids move in and out Transporters 44 |
-0.548 |
-0.539 |
| 227 |
259529_at |
AT1G12400
|
[Nucleotide excision repair, TFIIH, subunit TTDA] |
-0.545 |
-0.492 |
| 228 |
245736_at |
AT1G73330
|
ATDR4, drought-repressed 4, DR4, drought-repressed 4 |
-0.539 |
-0.389 |
| 229 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.536 |
0.087 |
| 230 |
255181_at |
AT4G08110
|
[CACTA-like transposase family (Ptta/En/Spm), has a 4.2e-66 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.533 |
-0.559 |
| 231 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.527 |
0.013 |
| 232 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.522 |
0.101 |
| 233 |
265048_at |
AT1G52050
|
[Mannose-binding lectin superfamily protein] |
-0.520 |
0.001 |
| 234 |
254327_at |
AT4G22490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.520 |
-0.264 |
| 235 |
247713_at |
AT5G59330
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.511 |
-0.188 |
| 236 |
267341_at |
AT2G44200
|
[CBF1-interacting co-repressor CIR, N-terminal] |
-0.510 |
-0.331 |
| 237 |
254724_at |
AT4G13630
|
unknown |
-0.508 |
-0.349 |
| 238 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
-0.506 |
-0.446 |
| 239 |
246194_at |
AT4G37000
|
ACD2, ACCELERATED CELL DEATH 2, ATRCCR, ARABIDOPSIS THALIANA RED CHLOROPHYLL CATABOLITE REDUCTASE |
-0.503 |
-0.024 |
| 240 |
255044_at |
AT4G09680
|
ATCTC1, CTC1, conserved telomere maintenance component 1 |
-0.501 |
-0.061 |
| 241 |
255260_at |
AT4G05040
|
[ankyrin repeat family protein] |
-0.498 |
0.007 |
| 242 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
-0.497 |
-0.493 |
| 243 |
256935_at |
AT3G22570
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.497 |
-0.081 |
| 244 |
265297_at |
AT2G14080
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.495 |
-0.240 |
| 245 |
257476_at |
AT1G80960
|
[F-box and Leucine Rich Repeat domains containing protein] |
-0.484 |
-0.372 |
| 246 |
267459_at |
AT2G33850
|
unknown |
-0.481 |
-0.796 |
| 247 |
262346_at |
AT1G63980
|
[D111/G-patch domain-containing protein] |
-0.478 |
0.044 |
| 248 |
260974_at |
AT1G53440
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.475 |
0.086 |
| 249 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.472 |
0.026 |
| 250 |
256352_at |
AT1G54970
|
ATPRP1, proline-rich protein 1, PRP1, proline-rich protein 1, RHS7, ROOT HAIR SPECIFIC 7 |
-0.470 |
0.084 |
| 251 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
-0.468 |
0.058 |
| 252 |
251165_at |
AT3G63340
|
[Protein phosphatase 2C family protein] |
-0.467 |
-0.491 |
| 253 |
248556_at |
AT5G50350
|
unknown |
-0.465 |
-0.023 |
| 254 |
256303_at |
AT1G69550
|
[disease resistance protein (TIR-NBS-LRR class)] |
-0.463 |
-0.415 |
| 255 |
255546_at |
AT4G01910
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.462 |
0.048 |
| 256 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
-0.453 |
-0.143 |
| 257 |
256920_at |
AT3G18980
|
ETP1, EIN2 targeting protein1 |
-0.450 |
-0.069 |
| 258 |
249331_at |
AT5G40950
|
RPL27, ribosomal protein large subunit 27 |
-0.449 |
-0.333 |
| 259 |
252593_at |
AT3G45590
|
ATSEN1, splicing endonuclease 1, SEN1, splicing endonuclease 1 |
-0.449 |
0.130 |
| 260 |
246393_at |
AT1G58150
|
unknown |
-0.447 |
-0.478 |
| 261 |
263873_at |
AT2G21860
|
[violaxanthin de-epoxidase-related] |
-0.446 |
-0.475 |
| 262 |
260012_at |
AT1G67865
|
unknown |
-0.444 |
-0.523 |
| 263 |
256021_at |
AT1G58270
|
ZW9 |
-0.439 |
-0.956 |
| 264 |
252833_at |
AT4G40090
|
AGP3, arabinogalactan protein 3 |
-0.437 |
-0.017 |
| 265 |
254326_at |
AT4G22610
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.436 |
-0.038 |
| 266 |
262637_at |
AT1G06640
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.434 |
-0.175 |
| 267 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
-0.432 |
-0.313 |
| 268 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.431 |
0.015 |
| 269 |
245966_at |
AT5G19790
|
RAP2.11, related to AP2 11 |
-0.425 |
-0.014 |
| 270 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.422 |
0.019 |
| 271 |
254084_at |
AT4G24940
|
AT-SAE1-1, ATSAE1A, SUMO-ACTIVATING ENZYME 1A, SAE1A, SUMO-activating enzyme 1A |
-0.420 |
0.051 |
| 272 |
254390_at |
AT4G21940
|
CPK15, calcium-dependent protein kinase 15 |
-0.420 |
0.097 |
| 273 |
258228_at |
AT3G27610
|
[Nucleotidylyl transferase superfamily protein] |
-0.418 |
-0.266 |
| 274 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
-0.416 |
-0.410 |
| 275 |
249522_at |
AT5G38700
|
unknown |
-0.413 |
-0.017 |
| 276 |
262750_at |
AT1G28710
|
[Nucleotide-diphospho-sugar transferase family protein] |
-0.413 |
-0.327 |
| 277 |
263883_at |
AT2G21830
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.413 |
-0.171 |
| 278 |
251226_at |
AT3G62680
|
ATPRP3, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 3, PRP3, proline-rich protein 3 |
-0.410 |
0.210 |
| 279 |
266384_at |
AT2G14660
|
unknown |
-0.406 |
-0.415 |
| 280 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.406 |
-0.119 |
| 281 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
-0.405 |
0.064 |
| 282 |
248212_at |
AT5G54020
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.405 |
0.038 |
| 283 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-0.398 |
0.184 |
| 284 |
250528_at |
AT5G08600
|
[U3 ribonucleoprotein (Utp) family protein] |
-0.397 |
-0.074 |
| 285 |
249612_at |
AT5G37290
|
[ARM repeat superfamily protein] |
-0.397 |
-0.037 |
| 286 |
252822_at |
AT4G39955
|
[alpha/beta-Hydrolases superfamily protein] |
-0.395 |
0.010 |
| 287 |
245055_at |
AT2G26470
|
unknown |
-0.395 |
-0.217 |
| 288 |
254914_at |
AT4G11290
|
[Peroxidase superfamily protein] |
-0.393 |
0.290 |
| 289 |
254044_at |
AT4G25820
|
ATXTH14, XTH14, xyloglucan endotransglucosylase/hydrolase 14, XTR9, xyloglucan endotransglycosylase 9 |
-0.389 |
0.184 |
| 290 |
248238_at |
AT5G53900
|
[Serine/threonine-protein kinase WNK (With No Lysine)-related] |
-0.389 |
-0.045 |
| 291 |
262719_at |
AT1G43590
|
unknown |
-0.387 |
-0.054 |
| 292 |
257460_at |
AT1G75580
|
SAUR51, SMALL AUXIN UPREGULATED RNA 51 |
-0.386 |
0.157 |
| 293 |
265102_at |
AT1G30870
|
[Peroxidase superfamily protein] |
-0.380 |
0.184 |
| 294 |
245106_at |
AT2G41650
|
unknown |
-0.378 |
0.077 |
| 295 |
248977_at |
AT5G45020
|
[Glutathione S-transferase family protein] |
-0.377 |
-0.377 |
| 296 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
-0.377 |
-0.349 |
| 297 |
250778_at |
AT5G05500
|
MOP10 |
-0.376 |
0.176 |
| 298 |
251600_at |
AT3G57710
|
[Protein kinase superfamily protein] |
-0.372 |
0.036 |
| 299 |
266276_at |
AT2G29330
|
TRI, tropinone reductase |
-0.367 |
0.024 |
| 300 |
265302_at |
AT2G13970
|
[Mutator-like transposase family, has a 4.1e-39 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
-0.367 |
-0.592 |
| 301 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
-0.365 |
-0.558 |
| 302 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
-0.364 |
0.123 |
| 303 |
264658_at |
AT1G09910
|
[Rhamnogalacturonate lyase family protein] |
-0.363 |
-0.126 |
| 304 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
-0.355 |
-0.034 |
| 305 |
254852_at |
AT4G12020
|
ATWRKY19, MAPKKK11, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 11, MEKK4, MAPK/ERK KINASE KINASE 4, WRKY19 |
-0.348 |
0.044 |
| 306 |
247345_at |
AT5G63760
|
ARI15, ARIADNE 15, ATARI15, ARABIDOPSIS ARIADNE 15 |
-0.346 |
-0.307 |
| 307 |
260758_at |
AT1G48930
|
AtGH9C1, glycosyl hydrolase 9C1, GH9C1, glycosyl hydrolase 9C1 |
-0.345 |
0.105 |
| 308 |
254965_at |
AT4G11090
|
TBL23, TRICHOME BIREFRINGENCE-LIKE 23 |
-0.345 |
0.024 |
| 309 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
-0.345 |
0.084 |
| 310 |
250923_at |
AT5G03455
|
ACR2, ARSENATE REDUCTASE 2, ARATH;CDC25, AtACR2, CDC25 |
-0.343 |
0.082 |
| 311 |
249978_at |
AT5G18850
|
unknown |
-0.343 |
-0.016 |
| 312 |
258962_at |
AT3G10570
|
CYP77A6, cytochrome P450, family 77, subfamily A, polypeptide 6 |
-0.342 |
-0.162 |
| 313 |
248976_at |
AT5G44930
|
ARAD2, ARABINAN DEFICIENT 2 |
-0.341 |
-0.142 |
| 314 |
255812_at |
AT4G10310
|
ATHKT1, HKT1, high-affinity K+ transporter 1, HKT1;1 |
-0.341 |
-0.166 |
| 315 |
252537_at |
AT3G45710
|
[Major facilitator superfamily protein] |
-0.340 |
-0.061 |
| 316 |
263970_at |
AT2G42850
|
CYP718, cytochrome P450, family 718 |
-0.338 |
0.010 |
| 317 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
-0.336 |
0.100 |
| 318 |
250755_at |
AT5G05750
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.336 |
-0.547 |
| 319 |
267645_at |
AT2G32860
|
BGLU33, beta glucosidase 33 |
-0.333 |
-0.025 |
| 320 |
246818_at |
AT5G27270
|
EMB976, EMBRYO DEFECTIVE 976 |
-0.332 |
-0.272 |
| 321 |
249996_at |
AT5G18600
|
[Thioredoxin superfamily protein] |
-0.330 |
-0.062 |
| 322 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
-0.329 |
-0.090 |
| 323 |
265439_at |
AT2G21045
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.328 |
0.223 |
| 324 |
247729_at |
AT5G59530
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.326 |
-0.018 |
| 325 |
245449_at |
AT4G16870
|
[copia-like retrotransposon family, has a 0. P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
-0.325 |
-0.653 |
| 326 |
263276_at |
AT2G14100
|
CYP705A13, cytochrome P450, family 705, subfamily A, polypeptide 13 |
-0.325 |
0.040 |
| 327 |
251360_at |
AT3G61210
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.324 |
-0.107 |
| 328 |
246374_at |
AT1G51840
|
[protein kinase-related] |
-0.320 |
0.075 |
| 329 |
246079_at |
AT5G20450
|
unknown |
-0.320 |
-0.120 |
| 330 |
255542_at |
AT4G01860
|
[Transducin family protein / WD-40 repeat family protein] |
-0.319 |
0.119 |
| 331 |
248879_at |
AT5G46180
|
DELTA-OAT, ornithine-delta-aminotransferase |
-0.319 |
0.021 |
| 332 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
-0.318 |
-0.034 |
| 333 |
256537_at |
AT1G33340
|
[ENTH/ANTH/VHS superfamily protein] |
-0.317 |
-0.131 |
| 334 |
249124_at |
AT5G43740
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.315 |
-0.618 |
| 335 |
263586_at |
AT2G25350
|
[Phox (PX) domain-containing protein] |
-0.315 |
-0.267 |
| 336 |
255814_at |
AT1G19900
|
[glyoxal oxidase-related protein] |
-0.314 |
-0.024 |
| 337 |
251728_at |
AT3G56300
|
[Cysteinyl-tRNA synthetase, class Ia family protein] |
-0.313 |
-0.092 |
| 338 |
252004_at |
AT3G52780
|
ATPAP20, PAP20 |
-0.311 |
-0.013 |
| 339 |
255934_at |
AT1G12740
|
CYP87A2, cytochrome P450, family 87, subfamily A, polypeptide 2 |
-0.310 |
-0.009 |
| 340 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
-0.309 |
0.060 |
| 341 |
266838_at |
AT2G25980
|
[Mannose-binding lectin superfamily protein] |
-0.306 |
0.023 |
| 342 |
251856_at |
AT3G54720
|
AMP1, ALTERED MERISTEM PROGRAM 1, AtAMP1, COP2, CONSTITUTIVE MORPHOGENESIS 2, HPT, HAUPTLING, MFO1, Multifolia, PT, PRIMORDIA TIMING |
-0.305 |
-0.132 |
| 343 |
254795_at |
AT4G12990
|
unknown |
-0.303 |
-0.379 |
| 344 |
258027_at |
AT3G19515
|
unknown |
-0.302 |
-0.002 |
| 345 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.302 |
-0.301 |
| 346 |
264454_at |
AT1G10320
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
-0.302 |
-0.082 |
| 347 |
245079_at |
AT2G23330
|
[copia-like retrotransposon family, has a 3.9e-195 P-value blast match to GB:AAA57005 Hopscotch polyprotein (Ty1_Copia-element) (Zea mays)] |
-0.302 |
-0.227 |
| 348 |
254313_at |
AT4G22460
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.301 |
-0.006 |
| 349 |
264790_at |
AT2G17820
|
AHK1, ATHK1, histidine kinase 1, HK1, histidine kinase 1 |
-0.301 |
-0.130 |
| 350 |
267189_at |
AT2G44180
|
MAP2A, methionine aminopeptidase 2A |
-0.301 |
-0.108 |
| 351 |
264835_at |
AT1G03550
|
AtSCAMP4, SCAMP4, Secretory carrier membrane protein 4 |
-0.301 |
-0.103 |
| 352 |
257772_at |
AT3G23080
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.300 |
-0.195 |
| 353 |
262748_at |
AT1G28610
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.298 |
0.074 |