|
probeID |
AGICode |
Annotation |
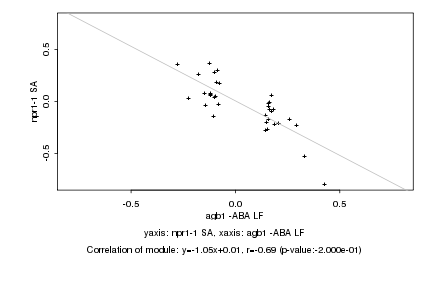
Log2 signal ratio
agb1 -ABA LF |
Log2 signal ratio
npr1-1 SA |
| 1 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
0.425 |
-0.789 |
| 2 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
0.329 |
-0.523 |
| 3 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
0.292 |
-0.225 |
| 4 |
255160_at |
AT4G07820
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.257 |
-0.171 |
| 5 |
250724_at |
AT5G06330
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.206 |
-0.209 |
| 6 |
245064_at |
AT2G39725
|
[LYR family of Fe/S cluster biogenesis protein] |
0.183 |
-0.213 |
| 7 |
252553_at |
AT3G45910
|
unknown |
0.178 |
-0.070 |
| 8 |
250426_at |
AT5G10510
|
AIL6, AINTEGUMENTA-like 6, PLT3, PLETHORA 3 |
0.172 |
0.067 |
| 9 |
266566_at |
AT2G24040
|
[Low temperature and salt responsive protein family] |
0.171 |
-0.092 |
| 10 |
258258_at |
AT3G26790
|
FUS3, FUSCA 3 |
0.162 |
-0.070 |
| 11 |
261936_at |
AT1G22600
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.162 |
-0.005 |
| 12 |
254327_at |
AT4G22490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.157 |
-0.011 |
| 13 |
263498_at |
AT2G42610
|
LSH10, LIGHT SENSITIVE HYPOCOTYLS 10 |
0.156 |
-0.040 |
| 14 |
250652_at |
AT5G06920
|
FLA21, FASCICLIN-like arabinogalactan protein 21 precursor |
0.155 |
-0.165 |
| 15 |
251366_at |
AT3G61340
|
[F-box and associated interaction domains-containing protein] |
0.150 |
-0.265 |
| 16 |
261700_at |
AT1G32690
|
unknown |
0.148 |
-0.195 |
| 17 |
252526_at |
AT3G46480
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.141 |
-0.276 |
| 18 |
265543_at |
AT2G28270
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.140 |
-0.128 |
| 19 |
260585_at |
AT2G43650
|
EMB2777, EMBRYO DEFECTIVE 2777 |
-0.281 |
0.361 |
| 20 |
261031_at |
AT1G17360
|
unknown |
-0.225 |
0.031 |
| 21 |
257207_at |
AT3G14900
|
EMB3120, EMBRYO DEFECTIVE 3120 |
-0.178 |
0.264 |
| 22 |
258997_at |
AT3G01810
|
unknown |
-0.151 |
0.082 |
| 23 |
255260_at |
AT4G05040
|
[ankyrin repeat family protein] |
-0.148 |
-0.036 |
| 24 |
251593_at |
AT3G57660
|
NRPA1, nuclear RNA polymerase A1 |
-0.128 |
0.375 |
| 25 |
246817_at |
AT5G27240
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.124 |
0.083 |
| 26 |
247686_at |
AT5G59700
|
[Protein kinase superfamily protein] |
-0.124 |
0.075 |
| 27 |
247185_at |
AT5G65460
|
KAC2, kinesin like protein for actin based chloroplast movement 2, KCA2, kinesin CDKA ;1 associated 2 |
-0.120 |
0.058 |
| 28 |
259380_at |
AT3G16340
|
ABCG29, ATP-binding cassette G29, AtABCG29, ATPDR1, PLEIOTROPIC DRUG RESISTANCE 1, PDR1, pleiotropic drug resistance 1 |
-0.108 |
-0.138 |
| 29 |
260404_at |
AT1G69950
|
[hAT-like transposase family (hobo/Ac/Tam3), has a 1.0e-71 P-value blast match to GB:CAA29005 ORFa of Maize Ac (hAT-element) (Zea mays)] |
-0.105 |
0.286 |
| 30 |
250569_at |
AT5G08130
|
BIM1 |
-0.101 |
0.043 |
| 31 |
249786_at |
AT5G24310
|
ABIL3, ABL interactor-like protein 3 |
-0.100 |
0.049 |
| 32 |
267110_at |
AT2G14800
|
unknown |
-0.093 |
0.185 |
| 33 |
264452_at |
AT1G10270
|
GRP23, glutamine-rich protein 23 |
-0.087 |
0.308 |
| 34 |
262547_at |
AT1G31270
|
unknown |
-0.083 |
-0.023 |
| 35 |
250256_at |
AT5G13650
|
SVR3, SUPPRESSOR OF VARIEGATION 3 |
-0.081 |
0.180 |