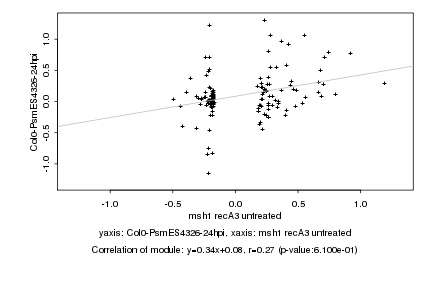
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
msh1 recA3 untreated |
Log2 signal ratio
Col0-PsmES4326-24hpi |
| 1 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
1.184 |
0.293 |
| 2 |
266752_at |
AT2G47000
|
ABCB4, ATP-binding cassette B4, AtABCB4, ATPGP4, ARABIDOPSIS P-GLYCOPROTEIN 4, MDR4, MULTIDRUG RESISTANCE 4, PGP4, P-GLYCOPROTEIN 4 |
0.918 |
0.776 |
| 3 |
250515_at |
AT5G09570
|
[Cox19-like CHCH family protein] |
0.796 |
0.114 |
| 4 |
250090_at |
AT5G17330
|
GAD, glutamate decarboxylase, GAD1, GLUTAMATE DECARBOXYLASE 1 |
0.740 |
0.800 |
| 5 |
263401_at |
AT2G04070
|
[MATE efflux family protein] |
0.711 |
0.706 |
| 6 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.698 |
0.275 |
| 7 |
266729_at |
AT2G03130
|
[Ribosomal protein L12/ ATP-dependent Clp protease adaptor protein ClpS family protein] |
0.686 |
0.087 |
| 8 |
247435_at |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
0.675 |
0.500 |
| 9 |
260522_x_at |
AT2G41730
|
unknown |
0.663 |
0.309 |
| 10 |
265422_at |
AT2G20800
|
NDB4, NAD(P)H dehydrogenase B4 |
0.661 |
0.158 |
| 11 |
254419_at |
AT4G21490
|
NDB3, NAD(P)H dehydrogenase B3 |
0.556 |
0.068 |
| 12 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.546 |
1.069 |
| 13 |
251432_at |
AT3G59820
|
AtLETM1, LETM1, leucine zipper-EF-hand-containing transmembrane protein 1 |
0.530 |
-0.031 |
| 14 |
265428_at |
AT2G20720
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.483 |
0.189 |
| 15 |
258934_at |
AT3G10140
|
RECA3, RECA homolog 3 |
0.478 |
-0.074 |
| 16 |
246584_at |
AT5G14730
|
unknown |
0.459 |
0.202 |
| 17 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.440 |
0.331 |
| 18 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.434 |
0.262 |
| 19 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.418 |
0.915 |
| 20 |
246282_at |
AT4G36580
|
[AAA-type ATPase family protein] |
0.404 |
-0.134 |
| 21 |
263515_at |
AT2G21640
|
unknown |
0.400 |
0.587 |
| 22 |
246461_at |
AT5G16930
|
[AAA-type ATPase family protein] |
0.399 |
-0.209 |
| 23 |
245879_at |
AT5G09420
|
AtmtOM64, ATTOC64-V, translocon at the outer membrane of chloroplasts 64-V, MTOM64, ARABIDOPSIS THALIANA TRANSLOCON AT THE OUTER MEMBRANE OF CHLOROPLASTS 64-V, OM64, outer membrane 64, TOC64-V, translocon at the outer membrane of chloroplasts 64-V |
0.367 |
0.190 |
| 24 |
265030_at |
AT1G61610
|
[S-locus lectin protein kinase family protein] |
0.360 |
0.973 |
| 25 |
262336_at |
AT1G64220
|
TOM7-2, translocase of outer membrane 7 kDa subunit 2 |
0.343 |
-0.018 |
| 26 |
257317_at |
ATMG01060
|
ORF107G |
0.339 |
0.006 |
| 27 |
252281_at |
AT3G49320
|
[Metal-dependent protein hydrolase] |
0.333 |
-0.092 |
| 28 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.323 |
0.547 |
| 29 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.317 |
0.027 |
| 30 |
247612_at |
AT5G60730
|
[Anion-transporting ATPase] |
0.291 |
-0.050 |
| 31 |
258397_at |
AT3G15357
|
unknown |
0.290 |
0.091 |
| 32 |
264868_at |
AT1G24095
|
[Putative thiol-disulphide oxidoreductase DCC] |
0.277 |
0.548 |
| 33 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.273 |
1.064 |
| 34 |
258879_at |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.268 |
0.089 |
| 35 |
247855_at |
AT5G58210
|
[hydroxyproline-rich glycoprotein family protein] |
0.268 |
0.284 |
| 36 |
258452_at |
AT3G22370
|
AOX1A, alternative oxidase 1A, ATAOX1A, AtHSR3, HSR3, hyper-sensitivity-related 3 |
0.263 |
0.387 |
| 37 |
257840_at |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
0.263 |
0.818 |
| 38 |
262491_at |
AT1G21650
|
SECA2 |
0.262 |
-0.126 |
| 39 |
259672_at |
AT1G68990
|
MGP3, male gametophyte defective 3 |
0.262 |
-0.252 |
| 40 |
258871_at |
AT3G03060
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.261 |
-0.022 |
| 41 |
260664_at |
AT1G19510
|
ATRL5, RAD-like 5, RL5, RAD-like 5, RSM4, RADIALIS-LIKE SANT/MYB 4 |
0.260 |
-0.253 |
| 42 |
265147_at |
AT1G51380
|
[DEA(D/H)-box RNA helicase family protein] |
0.259 |
-0.050 |
| 43 |
248101_at |
AT5G55200
|
MGE1, mitochondrial GrpE 1 |
0.251 |
0.285 |
| 44 |
262586_at |
AT1G15480
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.248 |
-0.222 |
| 45 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.242 |
0.189 |
| 46 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
0.241 |
0.175 |
| 47 |
244903_at |
ATMG00660
|
ORF149 |
0.236 |
0.177 |
| 48 |
248227_at |
AT5G53820
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.231 |
1.315 |
| 49 |
256416_at |
AT3G11050
|
ATFER2, ferritin 2, FER2, ferritin 2 |
0.225 |
0.196 |
| 50 |
250679_at |
AT5G06550
|
JMJ22, Jumonji domain-containing protein 22 |
0.225 |
-0.200 |
| 51 |
251455_at |
AT3G60100
|
CSY5, citrate synthase 5 |
0.224 |
0.155 |
| 52 |
249870_at |
AT5G23080
|
TGH, TOUGH |
0.215 |
0.117 |
| 53 |
244906_at |
ATMG00690
|
ORF240A |
0.212 |
0.037 |
| 54 |
267011_at |
AT2G39230
|
LOJ, LATERAL ORGAN JUNCTION |
0.210 |
0.232 |
| 55 |
251800_at |
AT3G55510
|
RBL, REBELOTE |
0.209 |
-0.435 |
| 56 |
264460_at |
AT1G10170
|
ATNFXL1, NF-X-like 1, NFXL1, NF-X-like 1 |
0.205 |
0.229 |
| 57 |
256107_at |
AT1G16830
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.205 |
-0.070 |
| 58 |
252864_at |
AT4G39740
|
HCC2, homologue of copper chaperone SCO1 2 |
0.203 |
0.041 |
| 59 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.202 |
0.302 |
| 60 |
252355_at |
AT3G48250
|
BIR6, Buthionine sulfoximine-insensitive roots 6 |
0.199 |
-0.321 |
| 61 |
251564_at |
AT3G58160
|
ATMYOS3, ATXIJ, MYOSIN XI J, MYA3, MYOSIN A3, XI-16, MYOSIN XI-16, XIJ |
0.195 |
-0.058 |
| 62 |
247989_at |
AT5G56350
|
[Pyruvate kinase family protein] |
0.194 |
0.372 |
| 63 |
255278_at |
AT4G04940
|
[transducin family protein / WD-40 repeat family protein] |
0.186 |
-0.357 |
| 64 |
257334_at |
ATMG01370
|
ORF111D |
0.185 |
-0.052 |
| 65 |
247676_at |
AT5G59980
|
AtRPP30, GAF1, GAMETOPHYTE DEFECTIVE 1, RPP30 |
0.177 |
-0.110 |
| 66 |
253013_at |
AT4G37910
|
mtHsc70-1, mitochondrial heat shock protein 70-1 |
0.177 |
-0.147 |
| 67 |
254778_at |
AT4G12750
|
[Homeodomain-like transcriptional regulator] |
0.177 |
-0.154 |
| 68 |
248228_at |
AT5G53800
|
unknown |
0.175 |
0.252 |
| 69 |
260948_at |
AT1G06100
|
[Fatty acid desaturase family protein] |
-0.501 |
0.043 |
| 70 |
258751_at |
AT3G05890
|
RCI2B, RARE-COLD-INDUCIBLE 2B |
-0.442 |
-0.064 |
| 71 |
262902_x_at |
AT1G59930
|
[MADS-box family protein] |
-0.429 |
-0.394 |
| 72 |
251284_at |
AT3G61840
|
unknown |
-0.393 |
0.157 |
| 73 |
261525_at |
AT1G14330
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.366 |
0.384 |
| 74 |
251842_at |
AT3G54580
|
[Proline-rich extensin-like family protein] |
-0.318 |
0.083 |
| 75 |
262566_at |
AT1G34310
|
ARF12, auxin response factor 12 |
-0.314 |
-0.432 |
| 76 |
260241_at |
AT1G63710
|
CYP86A7, cytochrome P450, family 86, subfamily A, polypeptide 7 |
-0.301 |
0.060 |
| 77 |
259987_at |
AT1G75030
|
ATLP-3, thaumatin-like protein 3, TLP-3, thaumatin-like protein 3 |
-0.282 |
-0.036 |
| 78 |
266211_at |
AT2G06820
|
unknown |
-0.279 |
0.053 |
| 79 |
260930_at |
AT1G02620
|
[Ras-related small GTP-binding family protein] |
-0.275 |
0.044 |
| 80 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.254 |
0.071 |
| 81 |
265937_at |
AT2G19610
|
[RING/U-box superfamily protein] |
-0.247 |
0.145 |
| 82 |
260642_at |
AT1G53260
|
[LOCATED IN: endomembrane system] |
-0.245 |
0.073 |
| 83 |
245444_at |
AT4G16740
|
ATTPS03, terpene synthase 03, TPS03, terpene synthase 03 |
-0.244 |
0.707 |
| 84 |
255140_x_at |
AT4G08410
|
[Proline-rich extensin-like family protein] |
-0.240 |
0.068 |
| 85 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
-0.236 |
0.431 |
| 86 |
247762_at |
AT5G59170
|
[Proline-rich extensin-like family protein] |
-0.235 |
-0.061 |
| 87 |
259578_at |
AT1G27990
|
unknown |
-0.227 |
0.012 |
| 88 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.226 |
-0.839 |
| 89 |
252833_at |
AT4G40090
|
AGP3, arabinogalactan protein 3 |
-0.223 |
-0.025 |
| 90 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
-0.222 |
-0.030 |
| 91 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.222 |
-0.750 |
| 92 |
247128_at |
AT5G66110
|
HIPP27, heavy metal associated isoprenylated plant protein 27 |
-0.222 |
-0.017 |
| 93 |
250327_at |
AT5G12050
|
unknown |
-0.217 |
-1.145 |
| 94 |
249375_at |
AT5G40730
|
AGP24, arabinogalactan protein 24, ATAGP24, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEIN 24 |
-0.217 |
0.489 |
| 95 |
252050_at |
AT3G52550
|
unknown |
-0.217 |
0.029 |
| 96 |
245794_at |
AT1G32170
|
XTH30, xyloglucan endotransglucosylase/hydrolase 30, XTR4, xyloglucan endotransglycosylase 4 |
-0.215 |
0.521 |
| 97 |
259235_at |
AT3G11600
|
unknown |
-0.214 |
0.711 |
| 98 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.209 |
-0.461 |
| 99 |
258005_at |
AT3G19390
|
[Granulin repeat cysteine protease family protein] |
-0.209 |
0.225 |
| 100 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
-0.209 |
1.223 |
| 101 |
255944_at |
AT5G28600
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT1G35770.1)] |
-0.209 |
0.022 |
| 102 |
265169_x_at |
AT1G23720
|
[Proline-rich extensin-like family protein] |
-0.208 |
0.030 |
| 103 |
256518_at |
AT1G66080
|
unknown |
-0.204 |
0.212 |
| 104 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
-0.203 |
-0.033 |
| 105 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.200 |
-0.213 |
| 106 |
253788_at |
AT4G28680
|
AtTYDC, TYDC, L-tyrosine decarboxylase, TYRDC, L-tyrosine decarboxylase, TYRDC1, L-TYROSINE DECARBOXYLASE 1 |
-0.200 |
0.028 |
| 107 |
250136_at |
AT5G15380
|
DRM1, domains rearranged methylase 1 |
-0.198 |
0.039 |
| 108 |
262734_at |
AT1G28640
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.198 |
0.092 |
| 109 |
264035_at |
AT2G03630
|
unknown |
-0.197 |
0.084 |
| 110 |
262416_at |
AT1G49390
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.196 |
0.108 |
| 111 |
262115_at |
AT1G02813
|
unknown |
-0.195 |
-0.004 |
| 112 |
253317_at |
AT4G33960
|
unknown |
-0.194 |
-0.065 |
| 113 |
252761_at |
AT3G42770
|
[F-box/RNI-like/FBD-like domains-containing protein] |
-0.193 |
-0.083 |
| 114 |
252118_at |
AT3G51400
|
unknown |
-0.191 |
0.103 |
| 115 |
256094_at |
AT1G20780
|
ATPUB44, ARABIDOPSIS THALIANA PLANT U-BOX 44, PUB44, PLANT U-BOX 44, SAUL1, senescence-associated E3 ubiquitin ligase 1 |
-0.190 |
0.162 |
| 116 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.189 |
-0.826 |
| 117 |
251857_at |
AT3G54770
|
ARP1, ABA-regulated RNA-binding protein 1 |
-0.188 |
0.007 |
| 118 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
-0.187 |
-0.216 |
| 119 |
260315_at |
AT1G63820
|
[CCT motif family protein] |
-0.185 |
0.135 |
| 120 |
254258_at |
AT4G23410
|
TET5, tetraspanin5 |
-0.184 |
-0.089 |
| 121 |
247110_at |
AT5G65830
|
ATRLP57, receptor like protein 57, RLP57, receptor like protein 57 |
-0.184 |
-0.150 |
| 122 |
255160_at |
AT4G07820
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.183 |
0.147 |
| 123 |
258540_at |
AT3G06990
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.183 |
0.085 |
| 124 |
258030_at |
AT3G27590
|
unknown |
-0.182 |
-0.038 |
| 125 |
262133_at |
AT1G78000
|
SEL1, SELENATE RESISTANT 1, SULTR1;2, sulfate transporter 1;2 |
-0.182 |
0.187 |
| 126 |
250808_at |
AT5G05150
|
ATATG18E, ARABIDOPSIS THALIANA HOMOLOG OF YEAST AUTOPHAGY 18E, ATG18E, autophagy-related gene 18E, G18E, autophagy-related gene 18E |
-0.181 |
0.041 |
| 127 |
266766_at |
AT2G46880
|
ATPAP14, PAP14, purple acid phosphatase 14 |
-0.181 |
0.119 |
| 128 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
-0.180 |
0.048 |
| 129 |
259590_at |
AT1G28160
|
[Integrase-type DNA-binding superfamily protein] |
-0.180 |
0.075 |
| 130 |
265157_at |
AT1G31030
|
[non-LTR retrotransposon family (LINE), has a 1.0e-41 P-value blast match to GB:NP_038603 L1 repeat, Tf subfamily, member 23 (LINE-element) (Mus musculus)] |
-0.178 |
0.053 |
| 131 |
259525_at |
AT1G12560
|
ATEXP7, ATEXPA7, expansin A7, ATHEXP ALPHA 1.26, EXP7, EXPA7, expansin A7 |
-0.177 |
-0.027 |
| 132 |
253454_at |
AT4G31875
|
unknown |
-0.177 |
0.098 |
| 133 |
248931_at |
AT5G46040
|
[Major facilitator superfamily protein] |
-0.177 |
-0.004 |
| 134 |
245040_at |
AT2G26520
|
unknown |
-0.176 |
-0.075 |
| 135 |
259589_at |
AT1G28135
|
unknown |
-0.174 |
-0.002 |