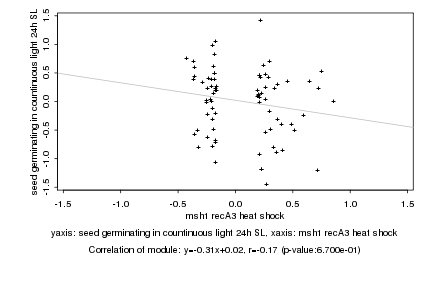
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
msh1 recA3 heat shock |
Log2 signal ratio
seed germinating in countinuous light 24h SL |
| 1 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.850 |
0.003 |
| 2 |
266752_at |
AT2G47000
|
ABCB4, ATP-binding cassette B4, AtABCB4, ATPGP4, ARABIDOPSIS P-GLYCOPROTEIN 4, MDR4, MULTIDRUG RESISTANCE 4, PGP4, P-GLYCOPROTEIN 4 |
0.742 |
0.542 |
| 3 |
250515_at |
AT5G09570
|
[Cox19-like CHCH family protein] |
0.716 |
0.238 |
| 4 |
265237_s_at |
ATMG00470
|
ORF122A |
0.711 |
-1.202 |
| 5 |
250090_at |
AT5G17330
|
GAD, glutamate decarboxylase, GAD1, GLUTAMATE DECARBOXYLASE 1 |
0.641 |
0.359 |
| 6 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.588 |
-0.232 |
| 7 |
256442_at |
AT3G10930
|
unknown |
0.511 |
-0.497 |
| 8 |
247435_at |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
0.487 |
-0.391 |
| 9 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.450 |
0.360 |
| 10 |
263401_at |
AT2G04070
|
[MATE efflux family protein] |
0.410 |
-0.849 |
| 11 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.393 |
-0.399 |
| 12 |
265428_at |
AT2G20720
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.361 |
-0.310 |
| 13 |
265422_at |
AT2G20800
|
NDB4, NAD(P)H dehydrogenase B4 |
0.361 |
0.298 |
| 14 |
257332_at |
ATMG01350
|
ORF145C |
0.351 |
-0.887 |
| 15 |
256416_at |
AT3G11050
|
ATFER2, ferritin 2, FER2, ferritin 2 |
0.336 |
0.228 |
| 16 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
0.325 |
-0.794 |
| 17 |
263061_at |
AT2G18190
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.301 |
-0.474 |
| 18 |
258934_at |
AT3G10140
|
RECA3, RECA homolog 3 |
0.296 |
0.705 |
| 19 |
254419_at |
AT4G21490
|
NDB3, NAD(P)H dehydrogenase B3 |
0.293 |
-0.165 |
| 20 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.284 |
0.427 |
| 21 |
252539_at |
AT3G45730
|
unknown |
0.267 |
-1.435 |
| 22 |
246461_at |
AT5G16930
|
[AAA-type ATPase family protein] |
0.260 |
0.482 |
| 23 |
244958_at |
ATMG00450
|
ORF106B |
0.259 |
-0.531 |
| 24 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.259 |
0.252 |
| 25 |
265242_at |
AT2G07705
|
unknown |
0.258 |
0.042 |
| 26 |
262459_at |
AT1G50400
|
[Eukaryotic porin family protein] |
0.243 |
0.639 |
| 27 |
257317_at |
ATMG01060
|
ORF107G |
0.226 |
-1.189 |
| 28 |
257336_at |
ATMG01410
|
ORF204, open reading frame 204 |
0.223 |
-1.176 |
| 29 |
262124_at |
AT1G59660
|
[Nucleoporin autopeptidase] |
0.223 |
0.146 |
| 30 |
257237_at |
AT3G14890
|
[phosphoesterase] |
0.218 |
0.435 |
| 31 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
0.212 |
1.428 |
| 32 |
263707_at |
AT1G09300
|
[Metallopeptidase M24 family protein] |
0.208 |
0.084 |
| 33 |
255193_at |
AT4G07400
|
VFB3, VIER F-box proteine 3 |
0.207 |
0.460 |
| 34 |
257401_at |
AT1G23550
|
SRO2, similar to RCD one 2 |
0.203 |
-0.017 |
| 35 |
260688_at |
AT1G17665
|
unknown |
0.202 |
-0.925 |
| 36 |
253019_at |
AT4G38010
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.201 |
0.461 |
| 37 |
262180_at |
AT1G78050
|
PGM, phosphoglycerate/bisphosphoglycerate mutase |
0.195 |
0.127 |
| 38 |
244903_at |
ATMG00660
|
ORF149 |
0.193 |
0.112 |
| 39 |
247707_at |
AT5G59450
|
[GRAS family transcription factor] |
0.192 |
0.196 |
| 40 |
262624_at |
AT1G06450
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
0.192 |
0.104 |
| 41 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.427 |
0.770 |
| 42 |
261069_at |
AT1G07410
|
ATRAB-A2B, ARABIDOPSIS RAB GTPASE HOMOLOG A2B, ATRABA2B, RAB GTPase homolog A2B, RAB-A2B, RAB GTPASE HOMOLOG A2B, RABA2b, RAB GTPase homolog A2B |
-0.374 |
0.387 |
| 43 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.374 |
0.713 |
| 44 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.365 |
-0.562 |
| 45 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.362 |
0.602 |
| 46 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.360 |
0.449 |
| 47 |
257670_at |
AT3G20340
|
unknown |
-0.333 |
-0.492 |
| 48 |
248015_at |
AT5G56370
|
[F-box/RNI-like/FBD-like domains-containing protein] |
-0.325 |
-0.791 |
| 49 |
246687_at |
AT5G33370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.295 |
0.349 |
| 50 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
-0.260 |
-0.010 |
| 51 |
259235_at |
AT3G11600
|
unknown |
-0.255 |
0.023 |
| 52 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
-0.249 |
0.231 |
| 53 |
261109_at |
AT1G75450
|
ATCKX5, ARABIDOPSIS THALIANA CYTOKININ OXIDASE 5, ATCKX6, CYTOKININ OXIDASE 6, CKX5, cytokinin oxidase 5 |
-0.248 |
-0.212 |
| 54 |
258177_at |
AT3G21660
|
[UBX domain-containing protein] |
-0.246 |
-0.616 |
| 55 |
258836_at |
AT3G07210
|
unknown |
-0.239 |
0.407 |
| 56 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
-0.219 |
0.047 |
| 57 |
260041_at |
AT1G68780
|
[RNI-like superfamily protein] |
-0.214 |
0.003 |
| 58 |
249895_at |
AT5G22500
|
FAR1, fatty acid reductase 1 |
-0.212 |
0.265 |
| 59 |
255878_at |
AT2G40620
|
[Basic-leucine zipper (bZIP) transcription factor family protein] |
-0.209 |
0.394 |
| 60 |
266836_at |
AT2G26000
|
BRIZ2, BRAP2 RING ZnF UBP domain-containing protein 2 |
-0.208 |
-0.106 |
| 61 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.206 |
-0.305 |
| 62 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
-0.204 |
-0.770 |
| 63 |
264007_at |
AT2G21140
|
ATPRP2, proline-rich protein 2, PRP2, proline-rich protein 2 |
-0.201 |
0.988 |
| 64 |
247457_at |
AT5G62170
|
TRM25, TON1 Recruiting Motif 25 |
-0.198 |
-0.481 |
| 65 |
260297_at |
AT1G80280
|
[alpha/beta-Hydrolases superfamily protein] |
-0.196 |
0.142 |
| 66 |
267555_at |
AT2G32765
|
ATSUMO5, SUM5, SUMO 5, SUMO5, small ubiquitinrelated modifier 5 |
-0.195 |
0.615 |
| 67 |
246395_at |
AT1G58170
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.191 |
0.396 |
| 68 |
249290_at |
AT5G41060
|
[DHHC-type zinc finger family protein] |
-0.190 |
0.506 |
| 69 |
259801_at |
AT1G72230
|
[Cupredoxin superfamily protein] |
-0.190 |
0.501 |
| 70 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
-0.187 |
0.835 |
| 71 |
245011_at |
ATCG00430
|
PSBG, photosystem II reaction center protein G |
-0.181 |
-0.700 |
| 72 |
254544_at |
AT4G19820
|
[Glycosyl hydrolase family protein with chitinase insertion domain] |
-0.181 |
-0.198 |
| 73 |
261012_at |
AT1G26600
|
CLE9, CLAVATA3/ESR-RELATED 9 |
-0.181 |
-1.052 |
| 74 |
259377_at |
AT3G16380
|
PAB6, poly(A) binding protein 6 |
-0.180 |
-0.666 |
| 75 |
249813_at |
AT5G23940
|
DCR, DEFECTIVE IN CUTICULAR RIDGES, EMB3009, EMBRYO DEFECTIVE 3009, PEL3, PERMEABLE LEAVES3 |
-0.180 |
1.065 |
| 76 |
252950_at |
AT4G38690
|
[PLC-like phosphodiesterases superfamily protein] |
-0.176 |
0.245 |
| 77 |
247048_at |
AT5G66560
|
[Phototropic-responsive NPH3 family protein] |
-0.176 |
0.198 |
| 78 |
247069_at |
AT5G66920
|
sks17, SKU5 similar 17 |
-0.173 |
0.279 |
| 79 |
256671_at |
AT3G52290
|
IQD3, IQ-domain 3 |
-0.172 |
0.206 |