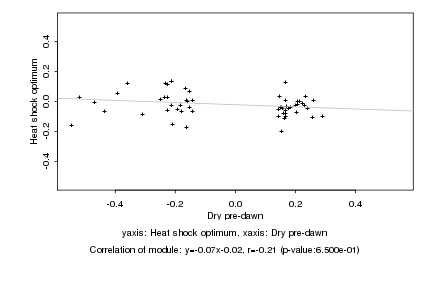
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Dry pre-dawn |
Log2 signal ratio
Heat shock optimum |
| 1 |
252090_at |
AT3G52130
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.287 |
-0.095 |
| 2 |
248321_at |
AT5G52740
|
[Copper transport protein family] |
0.257 |
0.011 |
| 3 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.254 |
-0.104 |
| 4 |
253036_at |
AT4G38340
|
NLP3, NIN-like protein 3 |
0.237 |
-0.045 |
| 5 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
0.230 |
0.037 |
| 6 |
247212_at |
AT5G65040
|
unknown |
0.227 |
-0.024 |
| 7 |
256481_at |
AT1G33430
|
[Galactosyltransferase family protein] |
0.223 |
-0.012 |
| 8 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
0.213 |
0.006 |
| 9 |
264341_at |
AT1G70270
|
unknown |
0.206 |
-0.016 |
| 10 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
0.206 |
0.003 |
| 11 |
248271_at |
AT5G53420
|
[CCT motif family protein] |
0.203 |
-0.073 |
| 12 |
250100_at |
AT5G16570
|
GLN1;4, glutamine synthetase 1;4 |
0.199 |
-0.026 |
| 13 |
253382_at |
AT4G33040
|
[Thioredoxin superfamily protein] |
0.183 |
-0.037 |
| 14 |
260261_at |
AT1G68450
|
PDE337, PIGMENT DEFECTIVE 337 |
0.175 |
-0.040 |
| 15 |
259766_at |
AT1G64360
|
unknown |
0.168 |
-0.033 |
| 16 |
245401_at |
AT4G17670
|
unknown |
0.166 |
-0.076 |
| 17 |
264590_at |
AT2G17710
|
unknown |
0.165 |
0.130 |
| 18 |
255310_at |
AT4G04955
|
ALN, allantoinase, ATALN, allantoinase |
0.165 |
0.009 |
| 19 |
251298_at |
AT3G62040
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.165 |
-0.056 |
| 20 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.164 |
-0.099 |
| 21 |
254174_at |
AT4G24120
|
ATYSL1, YELLOW STRIPE LIKE 1, YSL1, YELLOW STRIPE like 1 |
0.162 |
-0.109 |
| 22 |
248484_at |
AT5G51030
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.158 |
-0.076 |
| 23 |
257039_at |
AT3G19160
|
ATIPT8, ATP/ADP isopentenyltransferases, IPT8, ATP/ADP isopentenyltransferases, IPT8, isopentenyltransferase 8, PGA22 |
0.155 |
-0.040 |
| 24 |
263847_at |
AT2G36970
|
[UDP-Glycosyltransferase superfamily protein] |
0.151 |
-0.195 |
| 25 |
247727_at |
AT5G59490
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.151 |
-0.038 |
| 26 |
245133_at |
AT2G45310
|
GAE4, UDP-D-glucuronate 4-epimerase 4 |
0.147 |
-0.038 |
| 27 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
0.144 |
0.034 |
| 28 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
0.143 |
-0.047 |
| 29 |
255957_at |
AT1G22160
|
unknown |
0.141 |
-0.097 |
| 30 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
-0.548 |
-0.159 |
| 31 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.519 |
0.032 |
| 32 |
265387_at |
AT2G20670
|
unknown |
-0.470 |
-0.003 |
| 33 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
-0.437 |
-0.066 |
| 34 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
-0.393 |
0.056 |
| 35 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
-0.361 |
0.126 |
| 36 |
263805_at |
AT2G40400
|
unknown |
-0.311 |
-0.081 |
| 37 |
259015_at |
AT3G07350
|
unknown |
-0.252 |
0.016 |
| 38 |
245831_at |
AT1G48840
|
unknown |
-0.239 |
0.031 |
| 39 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
-0.236 |
0.127 |
| 40 |
258227_at |
AT3G15620
|
UVR3, UV REPAIR DEFECTIVE 3 |
-0.229 |
0.029 |
| 41 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-0.228 |
-0.058 |
| 42 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
-0.228 |
0.118 |
| 43 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
-0.215 |
0.135 |
| 44 |
262399_at |
AT1G49500
|
unknown |
-0.215 |
-0.022 |
| 45 |
245692_at |
AT5G04150
|
BHLH101 |
-0.215 |
0.138 |
| 46 |
256096_at |
AT1G13650
|
unknown |
-0.212 |
-0.151 |
| 47 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
-0.196 |
-0.048 |
| 48 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
-0.186 |
-0.020 |
| 49 |
249327_at |
AT5G40890
|
ATCLC-A, chloride channel A, ATCLCA, CLC-A, chloride channel A, CLC-A, CHLORIDE CHANNEL-A, CLCA, CHLORIDE CHANNEL A |
-0.182 |
-0.065 |
| 50 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-0.167 |
0.089 |
| 51 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
-0.165 |
0.007 |
| 52 |
266918_at |
AT2G45800
|
PLIM2a, PLIM2a |
-0.164 |
-0.170 |
| 53 |
253647_at |
AT4G29950
|
[Ypt/Rab-GAP domain of gyp1p superfamily protein] |
-0.162 |
0.006 |
| 54 |
255734_at |
AT1G25550
|
[myb-like transcription factor family protein] |
-0.154 |
-0.035 |
| 55 |
251356_at |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
-0.154 |
0.073 |
| 56 |
255878_at |
AT2G40620
|
[Basic-leucine zipper (bZIP) transcription factor family protein] |
-0.146 |
0.012 |
| 57 |
254469_at |
AT4G20470
|
unknown |
-0.144 |
-0.062 |