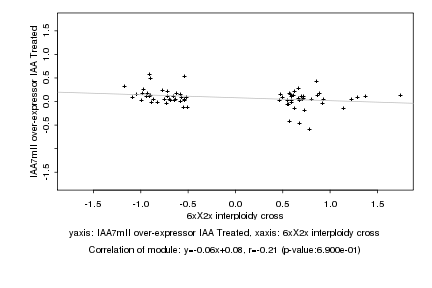
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
6xX2x interploidy cross |
Log2 signal ratio
IAA7mII over-expressor IAA Treated |
| 1 |
257944_at |
AT3G21850
|
ASK9, SKP1-like 9, SK9, SKP1-like 9 |
1.740 |
0.129 |
| 2 |
256497_at |
AT1G31580
|
CXC750, ECS1 |
1.370 |
0.108 |
| 3 |
261684_at |
AT1G47400
|
unknown |
1.289 |
0.103 |
| 4 |
262206_at |
AT2G01090
|
[Ubiquinol-cytochrome C reductase hinge protein] |
1.223 |
0.056 |
| 5 |
261848_at |
AT1G11590
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
1.134 |
-0.139 |
| 6 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
0.927 |
0.048 |
| 7 |
257945_at |
AT3G21860
|
ASK10, SKP1-like 10, SK10, SKP1-like 10 |
0.911 |
-0.038 |
| 8 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
0.882 |
0.183 |
| 9 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
0.867 |
0.140 |
| 10 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.849 |
0.429 |
| 11 |
245341_at |
AT4G16447
|
unknown |
0.799 |
0.053 |
| 12 |
256011_at |
AT1G19230
|
[Riboflavin synthase-like superfamily protein] |
0.773 |
-0.584 |
| 13 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.724 |
-0.176 |
| 14 |
253152_at |
AT4G35690
|
unknown |
0.719 |
0.073 |
| 15 |
246260_at |
AT1G31820
|
PUT1, POLYAMINE UPTAKE TRANSPORTER 1 |
0.713 |
0.109 |
| 16 |
259574_at |
AT1G35310
|
MLP168, MLP-like protein 168 |
0.699 |
0.051 |
| 17 |
261826_at |
AT1G11580
|
ATPMEPCRA, PMEPCRA, methylesterase PCR A |
0.688 |
0.114 |
| 18 |
254585_at |
AT4G19500
|
[nucleoside-triphosphatases] |
0.673 |
0.034 |
| 19 |
259606_at |
AT1G27920
|
MAP65-8, microtubule-associated protein 65-8 |
0.669 |
-0.454 |
| 20 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
0.664 |
0.082 |
| 21 |
252692_at |
AT3G43960
|
[Cysteine proteinases superfamily protein] |
0.659 |
0.288 |
| 22 |
261212_at |
AT1G12880
|
atnudt12, nudix hydrolase homolog 12, NUDT12, nudix hydrolase homolog 12 |
0.623 |
-0.142 |
| 23 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.621 |
0.224 |
| 24 |
250828_at |
AT5G05250
|
unknown |
0.604 |
0.147 |
| 25 |
252648_at |
AT3G44630
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.589 |
0.032 |
| 26 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
0.588 |
-0.012 |
| 27 |
245555_at |
AT4G15390
|
[HXXXD-type acyl-transferase family protein] |
0.586 |
0.122 |
| 28 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.577 |
0.157 |
| 29 |
247742_at |
AT5G58980
|
[Neutral/alkaline non-lysosomal ceramidase] |
0.569 |
-0.411 |
| 30 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
0.564 |
0.175 |
| 31 |
255484_at |
AT4G02540
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.555 |
-0.046 |
| 32 |
258434_at |
AT3G16770
|
ATEBP, ethylene-responsive element binding protein, EBP, ethylene-responsive element binding protein, ERF72, ETHYLENE RESPONSE FACTOR 72, RAP2.3, RELATED TO AP2 3 |
0.550 |
-0.048 |
| 33 |
252462_at |
AT3G47250
|
unknown |
0.547 |
0.026 |
| 34 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
0.496 |
0.094 |
| 35 |
263574_at |
AT2G16990
|
[Major facilitator superfamily protein] |
0.472 |
0.158 |
| 36 |
264958_at |
AT1G76960
|
unknown |
0.458 |
0.039 |
| 37 |
259045_at |
AT3G03410
|
[EF hand calcium-binding protein family] |
-1.178 |
0.334 |
| 38 |
260655_at |
AT1G19320
|
[Pathogenesis-related thaumatin superfamily protein] |
-1.090 |
0.104 |
| 39 |
267395_at |
AT2G44250
|
unknown |
-1.048 |
0.150 |
| 40 |
257681_at |
AT3G13370
|
unknown |
-1.003 |
0.042 |
| 41 |
250804_at |
AT5G05030
|
unknown |
-0.984 |
0.182 |
| 42 |
266386_at |
AT2G32370
|
HDG3, homeodomain GLABROUS 3 |
-0.974 |
0.268 |
| 43 |
265590_at |
AT2G20160
|
ASK17, ARABIDOPSIS SKP1-LIKE 17, MEO, MEIDOS |
-0.947 |
0.127 |
| 44 |
257123_at |
AT3G20030
|
[F-box and associated interaction domains-containing protein] |
-0.931 |
0.186 |
| 45 |
245305_at |
AT4G17215
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.918 |
0.579 |
| 46 |
250416_at |
AT5G11220
|
unknown |
-0.914 |
0.124 |
| 47 |
255608_at |
AT4G01140
|
unknown |
-0.905 |
0.504 |
| 48 |
246859_at |
AT5G25950
|
unknown |
-0.904 |
0.139 |
| 49 |
249479_at |
AT5G38960
|
[RmlC-like cupins superfamily protein] |
-0.892 |
-0.017 |
| 50 |
254310_at |
AT4G22430
|
[F-box family protein] |
-0.876 |
0.059 |
| 51 |
254450_at |
AT4G21080
|
AtDOF4.5, DOF4.5, DNA-binding with One Finger 4.5 |
-0.825 |
-0.002 |
| 52 |
261271_at |
AT1G26795
|
[Plant self-incompatibility protein S1 family] |
-0.781 |
0.241 |
| 53 |
250473_at |
AT5G10220
|
ANN6, annexin 6, ANNAT6, ANNEXIN ARABIDOPSIS THALIANA 6 |
-0.758 |
0.043 |
| 54 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
-0.736 |
-0.040 |
| 55 |
252767_at |
AT3G42830
|
[RING/U-box superfamily protein] |
-0.727 |
0.231 |
| 56 |
262327_at |
AT1G64130
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.721 |
0.116 |
| 57 |
258781_at |
AT3G11740
|
unknown |
-0.700 |
0.053 |
| 58 |
253839_at |
AT4G27890
|
[HSP20-like chaperones superfamily protein] |
-0.689 |
0.029 |
| 59 |
267345_at |
AT2G44240
|
unknown |
-0.658 |
0.107 |
| 60 |
249856_at |
AT5G22980
|
scpl47, serine carboxypeptidase-like 47 |
-0.651 |
0.032 |
| 61 |
267408_at |
AT2G34890
|
[CTP synthase family protein] |
-0.643 |
0.048 |
| 62 |
260437_at |
AT1G68380
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.625 |
0.188 |
| 63 |
253963_at |
AT4G26470
|
[Calcium-binding EF-hand family protein] |
-0.587 |
0.159 |
| 64 |
258707_at |
AT3G09480
|
[Histone superfamily protein] |
-0.582 |
0.018 |
| 65 |
249450_at |
AT5G39440
|
SnRK1.3, SNF1-related protein kinase 1.3 |
-0.570 |
0.088 |
| 66 |
260471_at |
AT1G11070
|
unknown |
-0.558 |
-0.124 |
| 67 |
261466_at |
AT1G07690
|
unknown |
-0.546 |
0.022 |
| 68 |
245511_at |
AT4G15755
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.546 |
0.550 |
| 69 |
250241_at |
AT5G13600
|
[Phototropic-responsive NPH3 family protein] |
-0.542 |
0.059 |
| 70 |
267585_s_at |
AT2G42090
|
ACT9, actin 9 |
-0.525 |
0.099 |
| 71 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
-0.517 |
-0.116 |