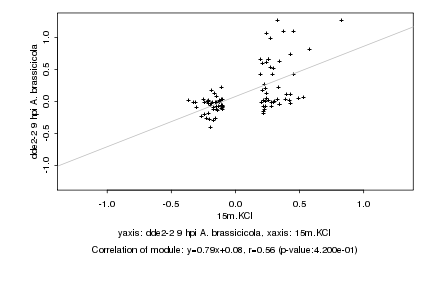
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
15m.KCl |
Log2 signal ratio
dde2-2 9 hpi A. brassicicola |
| 1 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.823 |
1.272 |
| 2 |
260522_x_at |
AT2G41730
|
unknown |
0.574 |
0.825 |
| 3 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.523 |
0.074 |
| 4 |
253505_at |
AT4G31970
|
CYP82C2, cytochrome P450, family 82, subfamily C, polypeptide 2 |
0.488 |
0.062 |
| 5 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
0.449 |
1.101 |
| 6 |
253643_at |
AT4G29780
|
unknown |
0.449 |
0.436 |
| 7 |
264912_at |
AT1G60750
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.427 |
0.117 |
| 8 |
250446_at |
AT5G10770
|
[Eukaryotic aspartyl protease family protein] |
0.425 |
-0.028 |
| 9 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.422 |
0.743 |
| 10 |
258939_at |
AT3G10020
|
unknown |
0.420 |
0.020 |
| 11 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
0.391 |
0.118 |
| 12 |
246250_at |
AT4G36880
|
CP1, cysteine proteinase1, RDL1, RD21A-LIKE PROTEASE1 |
0.388 |
0.036 |
| 13 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
0.367 |
1.104 |
| 14 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.342 |
-0.042 |
| 15 |
260406_at |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
0.340 |
0.631 |
| 16 |
254286_at |
AT4G22950
|
AGL19, AGAMOUS-like 19, GL19 |
0.337 |
-0.032 |
| 17 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.329 |
0.223 |
| 18 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
0.325 |
1.284 |
| 19 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.324 |
0.033 |
| 20 |
260851_at |
AT1G21890
|
UMAMIT19, Usually multiple acids move in and out Transporters 19 |
0.304 |
0.012 |
| 21 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.293 |
0.525 |
| 22 |
259729_at |
AT1G77640
|
[Integrase-type DNA-binding superfamily protein] |
0.292 |
-0.003 |
| 23 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.282 |
0.429 |
| 24 |
259017_at |
AT3G07310
|
unknown |
0.281 |
-0.069 |
| 25 |
244933_at |
ATCG01070
|
NDHE |
0.275 |
-0.008 |
| 26 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.273 |
0.547 |
| 27 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
0.272 |
0.987 |
| 28 |
255543_at |
AT4G01870
|
[tolB protein-related] |
0.255 |
0.670 |
| 29 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
0.253 |
0.018 |
| 30 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
0.250 |
0.668 |
| 31 |
255064_at |
AT4G08950
|
EXO, EXORDIUM |
0.242 |
0.051 |
| 32 |
261242_at |
AT1G32960
|
ATSBT3.3, SBT3.3 |
0.240 |
0.626 |
| 33 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
0.238 |
0.126 |
| 34 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.237 |
1.079 |
| 35 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
0.234 |
0.214 |
| 36 |
253259_at |
AT4G34410
|
RRTF1, redox responsive transcription factor 1 |
0.229 |
-0.066 |
| 37 |
259442_at |
AT1G02310
|
MAN1, endo-beta-mannanase 1 |
0.228 |
0.004 |
| 38 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
0.226 |
0.271 |
| 39 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
0.219 |
-0.114 |
| 40 |
254098_at |
AT4G25100
|
ATFSD1, ARABIDOPSIS FE SUPEROXIDE DISMUTASE 1, FSD1, Fe superoxide dismutase 1 |
0.217 |
-0.076 |
| 41 |
266611_at |
AT2G14960
|
GH3.1 |
0.217 |
0.027 |
| 42 |
266246_at |
AT2G27690
|
CYP94C1, cytochrome P450, family 94, subfamily C, polypeptide 1 |
0.213 |
-0.146 |
| 43 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
0.212 |
-0.182 |
| 44 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
0.210 |
0.600 |
| 45 |
249719_at |
AT5G35735
|
[Auxin-responsive family protein] |
0.204 |
0.185 |
| 46 |
257135_at |
AT3G12900
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.203 |
-0.005 |
| 47 |
247474_at |
AT5G62280
|
unknown |
0.192 |
0.436 |
| 48 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
0.190 |
0.672 |
| 49 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
-0.369 |
0.021 |
| 50 |
258285_at |
AT3G16140
|
PSAH-1, photosystem I subunit H-1 |
-0.330 |
-0.002 |
| 51 |
265287_at |
AT2G20260
|
PSAE-2, photosystem I subunit E-2 |
-0.318 |
-0.014 |
| 52 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
-0.311 |
-0.089 |
| 53 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-0.269 |
-0.222 |
| 54 |
245454_at |
AT4G16920
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.251 |
0.039 |
| 55 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.244 |
-0.189 |
| 56 |
254398_at |
AT4G21280
|
PSBQ, PHOTOSYSTEM II SUBUNIT Q, PSBQ-1, PHOTOSYSTEM II SUBUNIT Q-1, PSBQA, photosystem II subunit QA |
-0.243 |
-0.007 |
| 57 |
265083_at |
AT1G03820
|
unknown |
-0.228 |
-0.260 |
| 58 |
250165_at |
AT5G15290
|
CASP5, Casparian strip membrane domain protein 5 |
-0.224 |
0.001 |
| 59 |
246901_at |
AT5G25630
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.217 |
-0.182 |
| 60 |
245399_at |
AT4G17340
|
DELTA-TIP2, TIP2;2, tonoplast intrinsic protein 2;2 |
-0.214 |
0.023 |
| 61 |
266551_at |
AT2G35260
|
unknown |
-0.213 |
-0.018 |
| 62 |
252430_at |
AT3G47470
|
CAB4, LHCA4, light-harvesting chlorophyll-protein complex I subunit A4 |
-0.211 |
0.010 |
| 63 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
-0.206 |
-0.278 |
| 64 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
-0.198 |
-0.401 |
| 65 |
257117_at |
AT3G20160
|
[Terpenoid synthases superfamily protein] |
-0.195 |
-0.057 |
| 66 |
245277_at |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
-0.192 |
0.182 |
| 67 |
254102_at |
AT4G25050
|
ACP4, acyl carrier protein 4 |
-0.190 |
-0.025 |
| 68 |
265374_at |
AT2G06520
|
PSBX, photosystem II subunit X |
-0.181 |
-0.002 |
| 69 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.178 |
-0.287 |
| 70 |
254939_at |
AT4G10800
|
unknown |
-0.178 |
-0.116 |
| 71 |
266291_at |
AT2G29320
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.174 |
-0.082 |
| 72 |
265117_at |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.167 |
0.128 |
| 73 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
-0.163 |
-0.264 |
| 74 |
263436_at |
AT2G28690
|
unknown |
-0.161 |
-0.008 |
| 75 |
260300_at |
AT1G80340
|
ATGA3OX2, ARABIDOPSIS THALIANA GIBBERELLIN-3-OXIDASE 2, GA3OX2, gibberellin 3-oxidase 2, GA4H |
-0.153 |
-0.123 |
| 76 |
264815_at |
AT1G03620
|
[ELMO/CED-12 family protein] |
-0.152 |
-0.002 |
| 77 |
257311_at |
AT3G26570
|
ORF02, PHT2;1, phosphate transporter 2;1 |
-0.152 |
-0.067 |
| 78 |
258323_at |
AT3G22750
|
[Protein kinase superfamily protein] |
-0.150 |
0.094 |
| 79 |
256652_at |
AT3G18850
|
LPAT5, lysophosphatidyl acyltransferase 5 |
-0.148 |
-0.119 |
| 80 |
250550_at |
AT5G07870
|
[HXXXD-type acyl-transferase family protein] |
-0.143 |
-0.130 |
| 81 |
246847_at |
AT5G26820
|
ATIREG3, iron-regulated protein 3, IREG3, IRON REGULATED 3, IREG3, iron-regulated protein 3, MAR1, MULTIPLE ANTIBIOTIC RESISTANCE 1, RTS3 |
-0.136 |
-0.068 |
| 82 |
247679_at |
AT5G59540
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.132 |
0.029 |
| 83 |
255757_at |
AT4G00460
|
ATROPGEF3, ROPGEF3, ROP (rho of plants) guanine nucleotide exchange factor 3 |
-0.131 |
-0.008 |
| 84 |
260236_at |
AT1G74470
|
[Pyridine nucleotide-disulphide oxidoreductase family protein] |
-0.131 |
0.010 |
| 85 |
263137_at |
AT1G78660
|
ATGGH1, gamma-glutamyl hydrolase 1, GGH1, gamma-glutamyl hydrolase 1 |
-0.115 |
-0.095 |
| 86 |
257168_at |
AT3G24430
|
HCF101, HIGH-CHLOROPHYLL-FLUORESCENCE 101 |
-0.114 |
-0.054 |
| 87 |
262338_at |
AT1G64185
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
-0.113 |
-0.058 |
| 88 |
254606_at |
AT4G19030
|
AT-NLM1, ATNLM1, NOD26-LIKE MAJOR INTRINSIC PROTEIN 1, NIP1;1, NOD26-LIKE INTRINSIC PROTEIN 1;1, NLM1, NOD26-like major intrinsic protein 1 |
-0.110 |
0.042 |
| 89 |
257206_at |
AT3G16530
|
[Legume lectin family protein] |
-0.109 |
0.231 |
| 90 |
265819_at |
AT2G17972
|
unknown |
-0.108 |
-0.091 |
| 91 |
262916_at |
AT1G59700
|
ATGSTU16, glutathione S-transferase TAU 16, GSTU16, glutathione S-transferase TAU 16 |
-0.108 |
-0.064 |
| 92 |
258643_at |
AT3G08010
|
ATAB2 |
-0.107 |
-0.057 |
| 93 |
266931_at |
AT2G07730
|
[non-LTR retrotransposon family (LINE), has a 5.0e-32 P-value blast match to GB:NP_038603 L1 repeat, Tf subfamily, member 23 (LINE-element) (Mus musculus)] |
-0.107 |
0.044 |
| 94 |
261931_at |
AT1G22430
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.105 |
-0.118 |
| 95 |
261977_at |
AT1G37057
|
[hAT-like transposase family (hobo/Ac/Tam3), has a 7.8e-25 P-value blast match to GB:AAD24567 transposase Tag2 (hAT-element) (Arabidopsis thaliana)] |
-0.102 |
-0.065 |