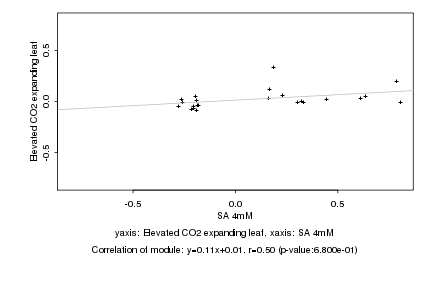
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
SA 4mM |
Log2 signal ratio
Elevated CO2 expanding leaf |
| 1 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
0.804 |
-0.005 |
| 2 |
264635_at |
AT1G65500
|
unknown |
0.782 |
0.199 |
| 3 |
256787_at |
AT3G13790
|
ATBFRUCT1, ATCWINV1, ARABIDOPSIS THALIANA CELL WALL INVERTASE 1 |
0.631 |
0.055 |
| 4 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.610 |
0.034 |
| 5 |
254665_at |
AT4G18340
|
[Glycosyl hydrolase superfamily protein] |
0.444 |
0.022 |
| 6 |
245181_at |
AT5G12420
|
[O-acyltransferase (WSD1-like) family protein] |
0.331 |
-0.002 |
| 7 |
256159_at |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
0.322 |
0.009 |
| 8 |
252882_at |
AT4G39675
|
unknown |
0.302 |
-0.007 |
| 9 |
254174_at |
AT4G24120
|
ATYSL1, YELLOW STRIPE LIKE 1, YSL1, YELLOW STRIPE like 1 |
0.227 |
0.068 |
| 10 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
0.185 |
0.334 |
| 11 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
0.165 |
0.119 |
| 12 |
253841_at |
AT4G27830
|
AtBGLU10, BGLU10, beta glucosidase 10 |
0.160 |
0.039 |
| 13 |
265383_at |
AT2G16780
|
MSI02, MSI2, MULTICOPY SUPPRESSOR OF IRA1 2, NFC02, NFC2, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP C 2 |
-0.279 |
-0.047 |
| 14 |
251886_at |
AT3G54260
|
TBL36, TRICHOME BIREFRINGENCE-LIKE 36 |
-0.266 |
0.029 |
| 15 |
246315_at |
AT3G56870
|
unknown |
-0.262 |
0.000 |
| 16 |
254728_at |
AT4G13690
|
unknown |
-0.216 |
-0.077 |
| 17 |
257181_at |
AT3G13190
|
unknown |
-0.206 |
-0.046 |
| 18 |
256784_at |
AT3G13674
|
unknown |
-0.206 |
-0.068 |
| 19 |
251354_at |
AT3G61090
|
[Putative endonuclease or glycosyl hydrolase] |
-0.197 |
0.057 |
| 20 |
253516_at |
AT4G31360
|
[selenium binding] |
-0.195 |
-0.081 |
| 21 |
259832_at |
AT1G69580
|
[Homeodomain-like superfamily protein] |
-0.192 |
0.014 |
| 22 |
250869_at |
AT5G03840
|
TFL-1, TERMINAL FLOWER 1, TFL1, TERMINAL FLOWER 1 |
-0.187 |
-0.030 |
| 23 |
254548_at |
AT4G19865
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.182 |
-0.031 |