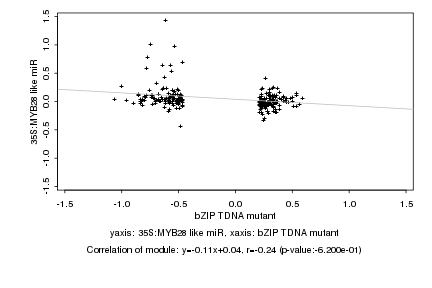
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
bZIP TDNA mutant |
Log2 signal ratio
35S:MYB28 like miR |
| 1 |
267431_at |
AT2G34870
|
MEE26, maternal effect embryo arrest 26 |
0.584 |
0.054 |
| 2 |
258742_at |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
0.561 |
-0.048 |
| 3 |
260181_at |
AT1G70710
|
ATGH9B1, glycosyl hydrolase 9B1, CEL1, CELLULASE 1, GH9B1, glycosyl hydrolase 9B1 |
0.532 |
-0.081 |
| 4 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
0.530 |
0.108 |
| 5 |
265031_at |
AT1G61590
|
[Protein kinase superfamily protein] |
0.530 |
0.156 |
| 6 |
263004_at |
AT1G54510
|
ATNEK1, NIMA-related serine/threonine kinase 1, NEK1, NIMA-related serine/threonine kinase 1 |
0.509 |
-0.076 |
| 7 |
253014_at |
AT4G37940
|
AGL21, AGAMOUS-like 21 |
0.500 |
0.001 |
| 8 |
266862_at |
AT2G26950
|
AtMYB104, myb domain protein 104, MYB104, myb domain protein 104 |
0.496 |
0.083 |
| 9 |
248368_at |
AT5G51950
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
0.484 |
0.056 |
| 10 |
265774_at |
AT2G07240
|
[cysteine-type peptidases] |
0.458 |
-0.015 |
| 11 |
253592_at |
AT4G30840
|
[Transducin/WD40 repeat-like superfamily protein] |
0.447 |
0.071 |
| 12 |
249461_at |
AT5G39640
|
[Putative endonuclease or glycosyl hydrolase] |
0.444 |
-0.004 |
| 13 |
265084_at |
AT1G03790
|
AtTZF4, SOM, SOMNUS, TZF4, Tandem CCCH Zinc Finger protein 4 |
0.437 |
0.030 |
| 14 |
250520_at |
AT5G08470
|
EMB2817, EMBRYO DEFECTIVE 2817, PEX1, peroxisome 1 |
0.420 |
0.099 |
| 15 |
256081_at |
AT1G20700
|
ATWOX14, WUSCHEL RELATED HOMEOBOX 14, WOX14, WUSCHEL related homeobox 14 |
0.420 |
0.005 |
| 16 |
257242_at |
AT3G24220
|
ATNCED6, NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 6, NCED6, nine-cis-epoxycarotenoid dioxygenase 6 |
0.416 |
0.082 |
| 17 |
253293_at |
AT4G33905
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
0.398 |
0.037 |
| 18 |
253516_at |
AT4G31360
|
[selenium binding] |
0.386 |
-0.054 |
| 19 |
266130_at |
AT2G44980
|
ASG3, ALTERED SEED GERMINATION 3, CHR10, chromatin remodeling 10 |
0.386 |
-0.125 |
| 20 |
246297_at |
AT3G51760
|
unknown |
0.383 |
0.057 |
| 21 |
259481_at |
AT1G18970
|
GLP4, germin-like protein 4 |
0.383 |
0.162 |
| 22 |
259573_at |
AT1G20390
|
[gypsy-like retrotransposon family, has a 1.5e-251 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
0.367 |
0.015 |
| 23 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
0.366 |
0.238 |
| 24 |
264253_at |
AT1G09170
|
[P-loop nucleoside triphosphate hydrolases superfamily protein with CH (Calponin Homology) domain] |
0.358 |
0.122 |
| 25 |
252253_at |
AT3G49300
|
[proline-rich family protein] |
0.357 |
-0.021 |
| 26 |
263257_at |
AT1G10520
|
AtPol{lambda}, Pol{lambda}, DNA polymerase {lambda} |
0.353 |
-0.191 |
| 27 |
263304_at |
AT2G01920
|
[ENTH/VHS/GAT family protein] |
0.353 |
0.015 |
| 28 |
258300_at |
AT3G23340
|
ckl10, casein kinase I-like 10 |
0.351 |
-0.137 |
| 29 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.349 |
-0.030 |
| 30 |
260341_at |
AT1G69170
|
AtSPL6, SPL6, SQUAMOSA PROMOTER BINDING PROTEIN (SBP)-domain transcription factor 6 |
0.347 |
-0.184 |
| 31 |
246047_at |
AT5G28870
|
[pseudogene] |
0.345 |
0.077 |
| 32 |
258522_at |
AT3G06660
|
[PAPA-1-like family protein / zinc finger (HIT type) family protein] |
0.336 |
0.118 |
| 33 |
264319_at |
AT1G04110
|
AtSDD1, SDD1, STOMATAL DENSITY AND DISTRIBUTION 1 |
0.333 |
0.264 |
| 34 |
257806_at |
AT3G18670
|
[Ankyrin repeat family protein] |
0.332 |
-0.075 |
| 35 |
250574_at |
AT5G08230
|
[Tudor/PWWP/MBT domain-containing protein] |
0.332 |
0.032 |
| 36 |
261547_at |
AT1G63500
|
BSK7, brassinosteroid-signaling kinase 7 |
0.331 |
-0.048 |
| 37 |
261591_at |
AT1G01740
|
BSK4, brassinosteroid-signaling kinase 4 |
0.331 |
-0.088 |
| 38 |
261829_at |
AT1G10680
|
ABCB10, ATP-binding cassette B10, PGP10, P-glycoprotein 10 |
0.331 |
0.082 |
| 39 |
256192_at |
AT1G30110
|
ATNUDX25, nudix hydrolase homolog 25, NUDX25, nudix hydrolase homolog 25 |
0.331 |
-0.159 |
| 40 |
260193_at |
AT1G67640
|
[Transmembrane amino acid transporter family protein] |
0.328 |
-0.013 |
| 41 |
267622_at |
AT2G39690
|
unknown |
0.328 |
0.097 |
| 42 |
247538_at |
AT5G61700
|
ABCA12, ATP-binding cassette A12, ATATH16, ARABIDOPSIS THALIANA ABC2 HOMOLOG 16, ATH16, ABC2 homolog 16 |
0.327 |
0.042 |
| 43 |
245237_at |
AT4G25520
|
SLK1, SEUSS-like 1 |
0.325 |
0.042 |
| 44 |
248815_at |
AT5G46920
|
[Intron maturase, type II family protein] |
0.323 |
0.026 |
| 45 |
266336_at |
AT2G32270
|
ZIP3, zinc transporter 3 precursor |
0.321 |
0.047 |
| 46 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
0.319 |
0.112 |
| 47 |
253626_at |
AT4G30640
|
[RNI-like superfamily protein] |
0.319 |
0.029 |
| 48 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
0.318 |
0.233 |
| 49 |
248495_at |
AT5G50780
|
[Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase family protein] |
0.316 |
-0.069 |
| 50 |
249440_at |
AT5G40030
|
[Protein kinase superfamily protein] |
0.306 |
0.029 |
| 51 |
253110_at |
AT4G35930
|
AtFBS4, FBS4, F-box stress induced 4 |
0.305 |
0.123 |
| 52 |
258289_at |
AT3G23450
|
unknown |
0.305 |
0.089 |
| 53 |
246816_at |
AT5G27230
|
[Frigida-like protein] |
0.304 |
0.102 |
| 54 |
258354_at |
AT3G14320
|
[Zinc finger, C3HC4 type (RING finger) family protein] |
0.300 |
-0.030 |
| 55 |
258422_at |
AT3G16710
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.297 |
-0.047 |
| 56 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
0.296 |
0.032 |
| 57 |
255900_at |
AT1G17830
|
unknown |
0.295 |
0.228 |
| 58 |
253966_at |
AT4G26520
|
AtFBA7, FBA7, fructose-bisphosphate aldolase 7 |
0.294 |
0.059 |
| 59 |
252482_at |
AT3G46670
|
UGT76E11, UDP-glucosyl transferase 76E11 |
0.292 |
0.168 |
| 60 |
247786_at |
AT5G58600
|
PMR5, POWDERY MILDEW RESISTANT 5, TBL44, TRICHOME BIREFRINGENCE-LIKE 44 |
0.291 |
0.006 |
| 61 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.291 |
0.116 |
| 62 |
264114_at |
AT2G31270
|
ATCDT1A, ARABIDOPSIS HOMOLOG OF YEAST CDT1 A, CDT1, ARABIDOPSIS HOMOLOG OF YEAST CDT1, CDT1A, homolog of yeast CDT1 A |
0.287 |
0.109 |
| 63 |
258798_at |
AT3G04540
|
[Cysteine-rich protein] |
0.286 |
-0.063 |
| 64 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
0.283 |
-0.197 |
| 65 |
254781_at |
AT4G12840
|
unknown |
0.281 |
-0.017 |
| 66 |
251756_at |
AT3G55820
|
[Fasciclin-like arabinogalactan family protein] |
0.279 |
-0.008 |
| 67 |
260628_at |
AT1G62320
|
[ERD (early-responsive to dehydration stress) family protein] |
0.279 |
-0.021 |
| 68 |
254206_at |
AT4G24180
|
ATTLP1, TLP1, THAUMATIN-LIKE PROTEIN 1 |
0.277 |
-0.176 |
| 69 |
246168_at |
AT5G32460
|
[Transcriptional factor B3 family protein] |
0.274 |
-0.174 |
| 70 |
247910_at |
AT5G57410
|
[Afadin/alpha-actinin-binding protein] |
0.272 |
-0.073 |
| 71 |
251580_at |
AT3G58450
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.271 |
-0.044 |
| 72 |
256653_at |
AT3G18870
|
[Mitochondrial transcription termination factor family protein] |
0.271 |
0.159 |
| 73 |
247086_at |
AT5G66320
|
GATA5, GATA transcription factor 5 |
0.271 |
-0.077 |
| 74 |
254286_at |
AT4G22950
|
AGL19, AGAMOUS-like 19, GL19 |
0.266 |
0.067 |
| 75 |
262534_at |
AT1G17040
|
ATSHA, ARABIDOPSIS THALIANA SH2 DOMAIN PROTEIN A, SHA, SH2 domain protein A, STATLA, STAT-TYPE LINKER-SH2 DOMAIN FACTOR A |
0.265 |
-0.075 |
| 76 |
256913_at |
AT3G23870
|
unknown |
0.262 |
-0.046 |
| 77 |
245551_at |
AT4G15350
|
CYP705A2, cytochrome P450, family 705, subfamily A, polypeptide 2 |
0.262 |
-0.006 |
| 78 |
267232_at |
AT2G44190
|
EDE1, ENDOSPERM DEFECTIVE 1, EMB3116, EMBRYO DEFECTIVE 3116, QWRF5, QWRF domain containing 5 |
0.260 |
-0.034 |
| 79 |
264355_at |
AT1G03210
|
[Phenazine biosynthesis PhzC/PhzF protein] |
0.257 |
-0.106 |
| 80 |
254317_at |
AT4G22510
|
unknown |
0.256 |
0.409 |
| 81 |
249491_at |
AT5G39130
|
[RmlC-like cupins superfamily protein] |
0.255 |
0.045 |
| 82 |
247725_at |
AT5G59410
|
[Rab5-interacting family protein] |
0.251 |
0.065 |
| 83 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
0.250 |
-0.299 |
| 84 |
246118_at |
AT5G20340
|
BG5, beta-1,3-glucanase 5 |
0.249 |
-0.076 |
| 85 |
260330_at |
AT1G80400
|
[RING/U-box superfamily protein] |
0.247 |
-0.011 |
| 86 |
264155_at |
AT1G65440
|
GTB1, global transcription factor group B1 |
0.244 |
-0.334 |
| 87 |
245579_at |
AT4G14810
|
unknown |
0.243 |
-0.070 |
| 88 |
256435_at |
AT3G11180
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.241 |
-0.028 |
| 89 |
249980_at |
AT5G18870
|
NSH5, nucleoside hydrolase 5 |
0.239 |
0.034 |
| 90 |
246256_at |
AT4G36770
|
[UDP-Glycosyltransferase superfamily protein] |
0.239 |
-0.049 |
| 91 |
264906_at |
AT2G17270
|
MPT1, mitochondrial phosphate transporter 1, PHT3;3, phosphate transporter 3;3 |
0.239 |
-0.061 |
| 92 |
254039_at |
AT4G25840
|
GPP1, glycerol-3-phosphatase 1 |
0.238 |
-0.000 |
| 93 |
261543_at |
AT1G63550
|
[Receptor-like protein kinase-related family protein] |
0.236 |
-0.039 |
| 94 |
257179_at |
AT3G13170
|
ATSPO11-1 |
0.234 |
-0.045 |
| 95 |
262767_at |
AT1G13170
|
ORP1D, OSBP(oxysterol binding protein)-related protein 1D |
0.233 |
0.030 |
| 96 |
248572_at |
AT5G49800
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.233 |
-0.219 |
| 97 |
259775_at |
AT1G29530
|
unknown |
0.232 |
0.239 |
| 98 |
253716_at |
AT4G29420
|
[F-box/RNI-like superfamily protein] |
0.230 |
0.029 |
| 99 |
248135_at |
AT5G54890
|
mCSF2, mitochondrial CAF-like splicing factor 2 |
0.229 |
-0.115 |
| 100 |
251670_at |
AT3G57190
|
PrfB3, peptide chain release factor 3 |
0.229 |
0.037 |
| 101 |
256396_at |
AT3G06150
|
unknown |
0.229 |
0.136 |
| 102 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
0.228 |
0.054 |
| 103 |
257245_at |
AT3G24110
|
[Calcium-binding EF-hand family protein] |
0.228 |
-0.046 |
| 104 |
262234_at |
AT1G48270
|
GCR1, G-protein-coupled receptor 1 |
0.227 |
-0.100 |
| 105 |
258409_at |
AT3G17640
|
[Leucine-rich repeat (LRR) family protein] |
0.226 |
0.222 |
| 106 |
254786_at |
AT4G12890
|
[Gamma interferon responsive lysosomal thiol (GILT) reductase family protein] |
0.225 |
0.043 |
| 107 |
258227_at |
AT3G15620
|
UVR3, UV REPAIR DEFECTIVE 3 |
0.224 |
-0.187 |
| 108 |
258551_at |
AT3G06890
|
unknown |
0.224 |
0.090 |
| 109 |
261849_at |
AT1G11370
|
[Pectin lyase-like superfamily protein] |
0.222 |
0.021 |
| 110 |
267032_at |
AT2G38490
|
CIPK22, CBL-interacting protein kinase 22, SnRK3.19, SNF1-RELATED PROTEIN KINASE 3.19 |
0.221 |
-0.074 |
| 111 |
256013_at |
AT1G19270
|
DA1, DA1 |
0.220 |
-0.045 |
| 112 |
248210_at |
AT5G54000
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.219 |
-0.077 |
| 113 |
256712_at |
AT2G34020
|
[Calcium-binding EF-hand family protein] |
0.219 |
0.005 |
| 114 |
263088_at |
AT2G16160
|
unknown |
0.217 |
-0.022 |
| 115 |
254817_at |
AT4G12460
|
ORP2B, OSBP(oxysterol binding protein)-related protein 2B |
0.217 |
-0.013 |
| 116 |
246257_at |
AT4G36690
|
ATU2AF65A |
0.213 |
-0.125 |
| 117 |
259973_at |
AT1G76630
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.211 |
-0.193 |
| 118 |
253893_at |
AT4G27390
|
unknown |
0.210 |
-0.066 |
| 119 |
262802_at |
AT1G20930
|
CDKB2;2, cyclin-dependent kinase B2;2 |
0.209 |
0.011 |
| 120 |
263417_at |
AT2G17180
|
DAZ1, DUO1-ACTIVATED ZINC FINGER 1 |
0.208 |
-0.052 |
| 121 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
0.207 |
0.075 |
| 122 |
255277_at |
AT4G04890
|
PDF2, protodermal factor 2 |
0.207 |
-0.012 |
| 123 |
267262_at |
AT2G22990
|
SCPL8, SERINE CARBOXYPEPTIDASE-LIKE 8, SNG1, sinapoylglucose 1 |
-1.072 |
0.037 |
| 124 |
264636_at |
AT1G65490
|
unknown |
-1.010 |
0.266 |
| 125 |
250243_at |
AT5G13630
|
ABAR, ABA-BINDING PROTEIN, CCH, CONDITIONAL CHLORINA, CCH1, CHLH, H SUBUNIT OF MG-CHELATASE, GUN5, GENOMES UNCOUPLED 5 |
-0.958 |
0.026 |
| 126 |
254225_at |
AT4G23670
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.898 |
-0.023 |
| 127 |
259106_at |
AT3G05490
|
RALFL22, ralf-like 22 |
-0.855 |
0.117 |
| 128 |
261488_at |
AT1G14345
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.855 |
0.131 |
| 129 |
256880_at |
AT3G26450
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.843 |
-0.031 |
| 130 |
245701_at |
AT5G04140
|
FD-GOGAT, FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE, GLS1, FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE 1, GLU1, glutamate synthase 1, GLUS |
-0.841 |
0.052 |
| 131 |
247025_at |
AT5G67030
|
ABA1, ABA DEFICIENT 1, ATABA1, ARABIDOPSIS THALIANA ABA DEFICIENT 1, ATZEP, ARABIDOPSIS THALIANA ZEAXANTHIN EPOXIDASE, IBS3, IMPAIRED IN BABA-INDUCED STERILITY 3, LOS6, LOW EXPRESSION OF OSMOTIC STRESS-RESPONSIVE GENES 6, NPQ2, NON-PHOTOCHEMICAL QUENCHING 2, ZEP, ZEAXANTHIN EPOXIDASE |
-0.840 |
0.015 |
| 132 |
257216_at |
AT3G14990
|
AtDJ1A, DJ-1 homolog A, DJ-1a, DJ1A, DJ-1 homolog A |
-0.827 |
0.013 |
| 133 |
263875_at |
AT2G21970
|
SEP2, stress enhanced protein 2 |
-0.820 |
0.045 |
| 134 |
263711_at |
AT2G20630
|
PIA1, PP2C induced by AVRRPM1 |
-0.818 |
-0.055 |
| 135 |
260014_at |
AT1G68010
|
ATHPR1, HPR, hydroxypyruvate reductase |
-0.806 |
0.019 |
| 136 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
-0.801 |
0.102 |
| 137 |
249482_at |
AT5G38980
|
unknown |
-0.794 |
0.072 |
| 138 |
259161_at |
AT3G01500
|
ATBCA1, BETA CARBONIC ANHYDRASE 1, ATSABP3, ARABIDOPSIS THALIANA SALICYLIC ACID-BINDING PROTEIN 3, CA1, carbonic anhydrase 1, SABP3, SALICYLIC ACID-BINDING PROTEIN 3 |
-0.790 |
0.119 |
| 139 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
-0.783 |
0.587 |
| 140 |
254098_at |
AT4G25100
|
ATFSD1, ARABIDOPSIS FE SUPEROXIDE DISMUTASE 1, FSD1, Fe superoxide dismutase 1 |
-0.779 |
0.792 |
| 141 |
252463_at |
AT3G47070
|
[LOCATED IN: thylakoid, chloroplast thylakoid membrane, chloroplast, chloroplast envelope] |
-0.762 |
0.211 |
| 142 |
245025_at |
ATCG00130
|
ATPF |
-0.751 |
1.019 |
| 143 |
256754_at |
AT3G25690
|
AtCHUP1, Arabidopsis thaliana CHLOROPLAST UNUSUAL POSITIONING 1, CHUP1, CHLOROPLAST UNUSUAL POSITIONING 1 |
-0.744 |
0.122 |
| 144 |
252037_at |
AT3G51920
|
ATCML9, CAM9, calmodulin 9, CML9, CALMODULIN LIKE PROTEIN 9 |
-0.741 |
0.086 |
| 145 |
248986_at |
AT5G45170
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.735 |
-0.047 |
| 146 |
246998_at |
AT5G67370
|
CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 |
-0.732 |
0.109 |
| 147 |
246554_at |
AT5G15450
|
APG6, ALBINO AND PALE GREEN 6, AtCLPB3, CLPB-P, CASEIN LYTIC PROTEINASE B-P, CLPB3, casein lytic proteinase B3 |
-0.729 |
0.070 |
| 148 |
249134_at |
AT5G43150
|
unknown |
-0.713 |
0.055 |
| 149 |
256636_at |
AT3G12000
|
[S-locus related protein SLR1, putative (S1)] |
-0.707 |
0.009 |
| 150 |
249244_at |
AT5G42270
|
FTSH5, VAR1, VARIEGATED 1 |
-0.704 |
-0.020 |
| 151 |
262499_at |
AT1G21770
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.700 |
0.019 |
| 152 |
264105_x_at |
AT2G13760
|
unknown |
-0.699 |
0.033 |
| 153 |
258736_at |
AT3G05900
|
[neurofilament protein-related] |
-0.699 |
0.329 |
| 154 |
254508_at |
AT4G20170
|
GALS3, galactan synthase 3 |
-0.685 |
0.128 |
| 155 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
-0.685 |
0.037 |
| 156 |
249010_at |
AT5G44580
|
unknown |
-0.681 |
0.134 |
| 157 |
247656_at |
AT5G59890
|
ADF4, actin depolymerizing factor 4, ATADF4 |
-0.675 |
0.012 |
| 158 |
261385_at |
AT1G05450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.666 |
0.008 |
| 159 |
265182_at |
AT1G23740
|
AOR, alkenal/one oxidoreductase |
-0.665 |
0.007 |
| 160 |
263759_at |
AT2G21290
|
unknown |
-0.658 |
0.074 |
| 161 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
-0.650 |
0.216 |
| 162 |
253197_at |
AT4G35250
|
HCF244, high chlorophyll fluorescence phenotype 244 |
-0.649 |
0.037 |
| 163 |
256603_at |
AT3G28270
|
unknown |
-0.643 |
0.649 |
| 164 |
254785_at |
AT4G12730
|
FLA2, FASCICLIN-like arabinogalactan 2 |
-0.639 |
0.242 |
| 165 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
-0.635 |
0.238 |
| 166 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
-0.630 |
-0.103 |
| 167 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.627 |
0.437 |
| 168 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.626 |
0.041 |
| 169 |
262483_at |
AT1G17220
|
FUG1, fu-gaeri1 |
-0.619 |
-0.034 |
| 170 |
247188_at |
AT5G65430
|
14-3-3KAPPA, 14-3-3 PROTEIN G-BOX FACTOR14 KAPPA, GF14 KAPPA, GRF8, general regulatory factor 8 |
-0.619 |
0.002 |
| 171 |
246896_at |
AT5G25550
|
[Leucine-rich repeat (LRR) family protein] |
-0.619 |
0.044 |
| 172 |
244969_at |
ATCG00650
|
RPS18, ribosomal protein S18 |
-0.618 |
1.448 |
| 173 |
259981_at |
AT1G76450
|
[Photosystem II reaction center PsbP family protein] |
-0.612 |
0.057 |
| 174 |
251218_at |
AT3G62410
|
CP12, CP12 DOMAIN-CONTAINING PROTEIN 1, CP12-2, CP12 domain-containing protein 2 |
-0.612 |
0.246 |
| 175 |
256241_at |
AT3G12390
|
[Nascent polypeptide-associated complex (NAC), alpha subunit family protein] |
-0.597 |
0.027 |
| 176 |
256516_at |
AT1G66150
|
TMK1, transmembrane kinase 1 |
-0.595 |
0.144 |
| 177 |
262304_at |
AT1G70890
|
MLP43, MLP-like protein 43 |
-0.591 |
-0.169 |
| 178 |
255248_at |
AT4G05180
|
PSBQ, PHOTOSYSTEM II SUBUNIT Q, PSBQ-2, photosystem II subunit Q-2, PSII-Q |
-0.586 |
0.006 |
| 179 |
250117_at |
AT5G16440
|
IPP1, isopentenyl diphosphate isomerase 1 |
-0.586 |
0.064 |
| 180 |
259804_at |
AT1G72160
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.585 |
0.085 |
| 181 |
249441_at |
AT5G39730
|
[AIG2-like (avirulence induced gene) family protein] |
-0.584 |
-0.024 |
| 182 |
255149_at |
AT4G08150
|
BP, BREVIPEDICELLUS, BP1, BREVIPEDICELLUS 1, KNAT1, KNOTTED-like from Arabidopsis thaliana |
-0.583 |
-0.137 |
| 183 |
245061_at |
AT2G39730
|
RCA, rubisco activase |
-0.582 |
-0.004 |
| 184 |
249710_at |
AT5G35630
|
ATGSL1, GLUTAMINE SYNTHETASE LIKE 1, GLN2, GLUTAMINE SYNTHETASE 2, GS2, glutamine synthetase 2 |
-0.579 |
0.010 |
| 185 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
-0.578 |
0.093 |
| 186 |
253264_at |
AT4G33950
|
ATOST1, OPEN STOMATA 1, OST1, OPEN STOMATA 1, P44, SNRK2-6, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-6, SNRK2.6, SNF1-RELATED PROTEIN KINASE 2.6, SRK2E |
-0.576 |
0.115 |
| 187 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
-0.575 |
0.077 |
| 188 |
244937_at |
ATCG01110
|
NDHH, NAD(P)H dehydrogenase subunit H |
-0.574 |
0.642 |
| 189 |
244972_at |
ATCG00680
|
PSBB, photosystem II reaction center protein B |
-0.568 |
0.535 |
| 190 |
258412_at |
AT3G17210
|
ATHS1, A. THALIANA HEAT STABLE PROTEIN 1, HS1, heat stable protein 1 |
-0.562 |
0.086 |
| 191 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
-0.561 |
0.119 |
| 192 |
256309_at |
AT1G30380
|
PSAK, photosystem I subunit K |
-0.558 |
0.007 |
| 193 |
246596_at |
AT5G14740
|
BETA CA2, BETA CARBONIC ANHYDRASE 2, CA18, CARBONIC ANHYDRASE 18, CA2, carbonic anhydrase 2 |
-0.558 |
0.094 |
| 194 |
260619_at |
AT1G08110
|
[lactoylglutathione lyase family protein / glyoxalase I family protein] |
-0.558 |
-0.018 |
| 195 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
-0.556 |
0.203 |
| 196 |
266566_at |
AT2G24040
|
[Low temperature and salt responsive protein family] |
-0.556 |
0.209 |
| 197 |
256525_at |
AT1G66180
|
[Eukaryotic aspartyl protease family protein] |
-0.552 |
0.042 |
| 198 |
256378_at |
AT1G66830
|
[Leucine-rich repeat protein kinase family protein] |
-0.551 |
0.126 |
| 199 |
250439_at |
AT5G10450
|
14-3-3lambda, 14-3-3 PROTEIN G-BOX FACTOR14 LAMBDA, AFT1, GRF6, G-box regulating factor 6 |
-0.548 |
-0.060 |
| 200 |
257066_at |
AT3G18280
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.546 |
0.140 |
| 201 |
245010_at |
ATCG00420
|
NDHJ, NADH dehydrogenase subunit J |
-0.540 |
0.983 |
| 202 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
-0.538 |
0.058 |
| 203 |
245047_at |
ATCG00020
|
PSBA, photosystem II reaction center protein A |
-0.536 |
0.182 |
| 204 |
257321_at |
ATMG01130
|
ORF106F |
-0.535 |
0.131 |
| 205 |
261746_at |
AT1G08380
|
PSAO, photosystem I subunit O |
-0.527 |
0.032 |
| 206 |
252929_at |
AT4G38970
|
AtFBA2, FBA2, fructose-bisphosphate aldolase 2 |
-0.526 |
0.017 |
| 207 |
260057_at |
AT1G78200
|
[Protein phosphatase 2C family protein] |
-0.525 |
-0.122 |
| 208 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
-0.519 |
-0.030 |
| 209 |
251519_at |
AT3G59400
|
GUN4, GENOMES UNCOUPLED 4 |
-0.514 |
0.132 |
| 210 |
259431_at |
AT1G01620
|
PIP1;3, PLASMA MEMBRANE INTRINSIC PROTEIN 1;3, PIP1C, plasma membrane intrinsic protein 1C, TMP-B |
-0.513 |
0.015 |
| 211 |
251121_at |
AT3G63420
|
AGG1, Ggamma-subunit 1, ATAGG1, GG1, Ggamma-subunit 1 |
-0.513 |
-0.011 |
| 212 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.510 |
0.213 |
| 213 |
245044_at |
AT2G26500
|
[cytochrome b6f complex subunit (petM), putative] |
-0.509 |
0.045 |
| 214 |
266123_at |
AT2G45180
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.509 |
0.199 |
| 215 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
-0.507 |
-0.051 |
| 216 |
251962_at |
AT3G53420
|
AtPIP2;1, PIP2, PLASMA MEMBRANE INTRINSIC PROTEIN 2, PIP2;1, PLASMA MEMBRANE INTRINSIC PROTEIN 2;1, PIP2A, plasma membrane intrinsic protein 2A |
-0.504 |
-0.028 |
| 217 |
260481_at |
AT1G10960
|
ATFD1, ferredoxin 1, FD1, ferredoxin 1 |
-0.501 |
0.054 |
| 218 |
266716_at |
AT2G46820
|
CURT1B, CURVATURE THYLAKOID 1B, PSAP, PSI-P, photosystem I P subunit, PTAC8, PLASTID TRANSCRIPTIONALLY ACTIVE 8, TMP14, THYLAKOID MEMBRANE PHOSPHOPROTEIN OF 14 KDA |
-0.500 |
0.017 |
| 219 |
257807_at |
AT3G26650
|
GAPA, glyceraldehyde 3-phosphate dehydrogenase A subunit, GAPA-1, GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE A SUBUNIT 1 |
-0.499 |
0.018 |
| 220 |
259803_at |
AT1G72150
|
PATL1, PATELLIN 1 |
-0.498 |
0.102 |
| 221 |
257896_at |
AT3G16920
|
ATCTL2, CTL2, chitinase-like protein 2 |
-0.495 |
-0.118 |
| 222 |
250498_at |
AT5G09660
|
PMDH2, peroxisomal NAD-malate dehydrogenase 2 |
-0.494 |
0.007 |
| 223 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
-0.489 |
0.127 |
| 224 |
264185_at |
AT1G54780
|
AtTLP18.3, TLP18.3, thylakoid lumen protein 18.3 |
-0.488 |
0.027 |
| 225 |
260025_at |
AT1G30070
|
[SGS domain-containing protein] |
-0.487 |
0.112 |
| 226 |
263709_at |
AT1G09310
|
unknown |
-0.486 |
0.039 |
| 227 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
-0.486 |
0.077 |
| 228 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.486 |
-0.428 |
| 229 |
263350_at |
AT2G13360
|
AGT, alanine:glyoxylate aminotransferase, AGT1, ALANINE:GLYOXYLATE AMINOTRANSFERASE 1, SGAT, L-serine:glyoxylate aminotransferase |
-0.485 |
0.038 |
| 230 |
252255_at |
AT3G49220
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.485 |
0.042 |
| 231 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.483 |
0.023 |
| 232 |
259727_at |
AT1G60950
|
ATFD2, FERREDOXIN 2, FD2, FED A |
-0.482 |
0.019 |
| 233 |
263223_at |
AT1G30630
|
[Coatomer epsilon subunit] |
-0.481 |
0.041 |
| 234 |
247882_at |
AT5G57785
|
unknown |
-0.481 |
0.115 |
| 235 |
265247_at |
AT2G43030
|
[Ribosomal protein L3 family protein] |
-0.481 |
0.018 |
| 236 |
262800_at |
AT1G20960
|
emb1507, embryo defective 1507 |
-0.479 |
0.049 |
| 237 |
263537_at |
AT2G24790
|
ATCOL3, BBX4, B-box domain protein 4, COL3, CONSTANS-like 3 |
-0.478 |
0.026 |
| 238 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.472 |
-0.085 |
| 239 |
249978_at |
AT5G18850
|
unknown |
-0.470 |
0.019 |
| 240 |
261072_at |
AT1G07340
|
ATSTP2, sugar transporter 2, STP2, sugar transporter 2 |
-0.470 |
0.045 |
| 241 |
246270_at |
AT4G36500
|
unknown |
-0.468 |
-0.068 |
| 242 |
244933_at |
ATCG01070
|
NDHE |
-0.468 |
0.702 |
| 243 |
263676_at |
AT1G09340
|
CRB, chloroplast RNA binding, CSP41B, CHLOROPLAST STEM-LOOP BINDING PROTEIN OF 41 KDA, HIP1.3, heteroglycan-interacting protein 1.3 |
-0.467 |
-0.008 |