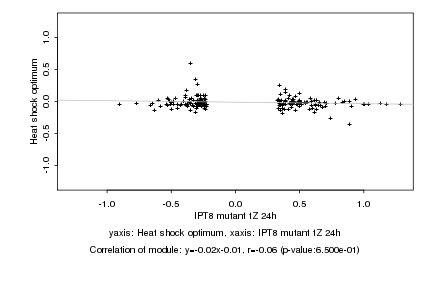
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
IPT8 mutant tZ 24h |
Log2 signal ratio
Heat shock optimum |
| 1 |
256631_at |
AT3G28320
|
unknown |
1.283 |
-0.032 |
| 2 |
259388_at |
AT1G13420
|
ATST4B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 4B, ST4B, sulfotransferase 4B |
1.173 |
-0.032 |
| 3 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
1.125 |
-0.028 |
| 4 |
248729_at |
AT5G48010
|
AtTHAS1, Arabidopsis thaliana thalianol synthase 1, THAS, THALIANOL SYNTHASE, THAS1, thalianol synthase 1 |
1.029 |
-0.034 |
| 5 |
253164_at |
AT4G35725
|
unknown |
1.000 |
-0.040 |
| 6 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
0.995 |
-0.032 |
| 7 |
254907_at |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.929 |
0.047 |
| 8 |
249599_at |
AT5G37990
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.904 |
-0.075 |
| 9 |
266078_at |
AT2G40670
|
ARR16, response regulator 16, RR16, response regulator 16 |
0.886 |
-0.347 |
| 10 |
247956_at |
AT5G56970
|
ATCKX3, CKX3, cytokinin oxidase 3 |
0.881 |
0.015 |
| 11 |
264404_at |
AT2G25160
|
CYP82F1, cytochrome P450, family 82, subfamily F, polypeptide 1 |
0.849 |
0.006 |
| 12 |
262608_at |
AT1G14120
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.832 |
0.000 |
| 13 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
0.796 |
0.062 |
| 14 |
256602_at |
AT3G28310
|
unknown |
0.777 |
-0.016 |
| 15 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
0.734 |
-0.253 |
| 16 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
0.709 |
-0.020 |
| 17 |
256368_at |
AT1G66800
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.707 |
-0.019 |
| 18 |
252537_at |
AT3G45710
|
[Major facilitator superfamily protein] |
0.701 |
-0.071 |
| 19 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
0.688 |
-0.007 |
| 20 |
262915_at |
AT1G59940
|
ARR3, response regulator 3 |
0.678 |
-0.080 |
| 21 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
0.658 |
-0.059 |
| 22 |
254632_at |
AT4G18630
|
unknown |
0.650 |
-0.012 |
| 23 |
259846_at |
AT1G72140
|
[Major facilitator superfamily protein] |
0.645 |
-0.061 |
| 24 |
248432_at |
AT5G51390
|
unknown |
0.629 |
0.028 |
| 25 |
267012_at |
AT2G39220
|
PLA IIB, PLP6, PATATIN-like protein 6 |
0.628 |
-0.116 |
| 26 |
257467_at |
AT1G31320
|
LBD4, LOB domain-containing protein 4 |
0.625 |
0.016 |
| 27 |
246251_at |
AT4G37220
|
[Cold acclimation protein WCOR413 family] |
0.619 |
-0.060 |
| 28 |
255385_at |
AT4G03610
|
[Metallo-hydrolase/oxidoreductase superfamily protein] |
0.616 |
-0.033 |
| 29 |
252374_at |
AT3G48100
|
ARR5, response regulator 5, ATRR2, ARABIDOPSIS THALIANA RESPONSE REGULATOR 2, IBC6, INDUCED BY CYTOKININ 6, RR5, response regulator 5 |
0.612 |
-0.161 |
| 30 |
252639_at |
AT3G44550
|
FAR5, fatty acid reductase 5 |
0.611 |
0.020 |
| 31 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
0.594 |
-0.105 |
| 32 |
247754_at |
AT5G59080
|
unknown |
0.592 |
0.005 |
| 33 |
251992_at |
AT3G53350
|
MIDD1, Microtubule Depletion Domain 1, RIP3, ROP interactive partner 3, RIP4, ROP interactive partner 4 |
0.588 |
-0.055 |
| 34 |
265083_at |
AT1G03820
|
unknown |
0.580 |
0.061 |
| 35 |
264790_at |
AT2G17820
|
AHK1, ATHK1, histidine kinase 1, HK1, histidine kinase 1 |
0.571 |
-0.111 |
| 36 |
248723_at |
AT5G47950
|
[HXXXD-type acyl-transferase family protein] |
0.561 |
-0.015 |
| 37 |
247097_at |
AT5G66460
|
AtMAN7, MAN7, endo-beta-mannase 7 |
0.551 |
-0.005 |
| 38 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.544 |
-0.026 |
| 39 |
253774_at |
AT4G28530
|
anac074, NAC domain containing protein 74, NAC074, NAC domain containing protein 74 |
0.536 |
-0.012 |
| 40 |
252202_at |
AT3G50300
|
[HXXXD-type acyl-transferase family protein] |
0.514 |
-0.038 |
| 41 |
254064_at |
AT4G25410
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.513 |
-0.015 |
| 42 |
263737_at |
AT1G60010
|
unknown |
0.511 |
-0.045 |
| 43 |
264138_at |
AT1G78950
|
AtBAS, AtLUP4, BAS, beta-amyrin synthase |
0.500 |
0.000 |
| 44 |
250327_at |
AT5G12050
|
unknown |
0.498 |
-0.076 |
| 45 |
248732_at |
AT5G48070
|
ATXTH20, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 20, XTH20, xyloglucan endotransglucosylase/hydrolase 20 |
0.495 |
-0.004 |
| 46 |
259653_at |
AT1G55240
|
unknown |
0.495 |
-0.038 |
| 47 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
0.492 |
0.139 |
| 48 |
252433_at |
AT3G47560
|
[alpha/beta-Hydrolases superfamily protein] |
0.492 |
0.021 |
| 49 |
258938_at |
AT3G10080
|
[RmlC-like cupins superfamily protein] |
0.489 |
-0.017 |
| 50 |
260053_at |
AT1G78120
|
TPR12, tetratricopeptide repeat 12 |
0.489 |
0.009 |
| 51 |
252536_at |
AT3G45700
|
[Major facilitator superfamily protein] |
0.482 |
-0.034 |
| 52 |
262801_at |
AT1G21010
|
unknown |
0.472 |
-0.045 |
| 53 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
0.464 |
0.094 |
| 54 |
266372_at |
AT2G41310
|
ARR8, RESPONSE REGULATOR 8, ATRR3, response regulator 3, RR3, response regulator 3 |
0.463 |
-0.135 |
| 55 |
262704_at |
AT1G16530
|
ASL9, ASYMMETRIC LEAVES 2-like 9, LBD3, LOB DOMAIN-CONTAINING PROTEIN 3 |
0.454 |
-0.012 |
| 56 |
266418_at |
AT2G38750
|
ANNAT4, annexin 4, AtANN4 |
0.452 |
0.007 |
| 57 |
248423_at |
AT5G51670
|
unknown |
0.451 |
-0.010 |
| 58 |
260034_at |
AT1G68810
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.448 |
0.046 |
| 59 |
263380_at |
AT2G40200
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.446 |
0.012 |
| 60 |
256447_at |
AT1G33440
|
AtNPF4.4, NPF4.4, NRT1/ PTR family 4.4 |
0.443 |
0.041 |
| 61 |
264071_at |
AT2G27920
|
SCPL51, serine carboxypeptidase-like 51 |
0.441 |
-0.038 |
| 62 |
250860_at |
AT5G04770
|
ATCAT6, ARABIDOPSIS THALIANA CATIONIC AMINO ACID TRANSPORTER 6, CAT6, cationic amino acid transporter 6 |
0.434 |
-0.024 |
| 63 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.430 |
0.009 |
| 64 |
253042_at |
AT4G37550
|
[Acetamidase/Formamidase family protein] |
0.430 |
-0.085 |
| 65 |
250200_at |
AT5G14130
|
[Peroxidase superfamily protein] |
0.428 |
-0.036 |
| 66 |
247228_at |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
0.417 |
-0.030 |
| 67 |
247360_at |
AT5G63450
|
CYP94B1, cytochrome P450, family 94, subfamily B, polypeptide 1 |
0.416 |
0.105 |
| 68 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
0.412 |
-0.112 |
| 69 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
0.408 |
0.059 |
| 70 |
261934_at |
AT1G22400
|
ATUGT85A1, ARABIDOPSIS THALIANA UDP-GLUCOSYL TRANSFERASE 85A1, UGT85A1 |
0.389 |
0.017 |
| 71 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
0.387 |
-0.044 |
| 72 |
261109_at |
AT1G75450
|
ATCKX5, ARABIDOPSIS THALIANA CYTOKININ OXIDASE 5, ATCKX6, CYTOKININ OXIDASE 6, CKX5, cytokinin oxidase 5 |
0.387 |
0.002 |
| 73 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.386 |
0.149 |
| 74 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
0.384 |
0.191 |
| 75 |
262940_at |
AT1G79520
|
[Cation efflux family protein] |
0.382 |
-0.124 |
| 76 |
258342_at |
AT3G22800
|
[Leucine-rich repeat (LRR) family protein] |
0.373 |
-0.020 |
| 77 |
245439_at |
AT4G16670
|
unknown |
0.368 |
-0.020 |
| 78 |
267637_at |
AT2G42190
|
unknown |
0.366 |
-0.181 |
| 79 |
247005_at |
AT5G67520
|
adenosine-5'-phosphosulfate (APS) kinase 4 |
0.365 |
-0.056 |
| 80 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
0.359 |
-0.100 |
| 81 |
267590_at |
AT2G39700
|
ATEXP4, ATEXPA4, expansin A4, ATHEXP ALPHA 1.6, EXPA4, expansin A4 |
0.352 |
0.017 |
| 82 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
0.351 |
-0.050 |
| 83 |
256713_at |
AT2G34060
|
[Peroxidase superfamily protein] |
0.349 |
-0.131 |
| 84 |
262785_at |
AT1G10750
|
unknown |
0.348 |
-0.032 |
| 85 |
251665_at |
AT3G57040
|
ARR9, response regulator 9, ATRR4, RESPONSE REGULATOR 4 |
0.347 |
-0.128 |
| 86 |
251886_at |
AT3G54260
|
TBL36, TRICHOME BIREFRINGENCE-LIKE 36 |
0.345 |
0.110 |
| 87 |
248674_at |
AT5G48800
|
[Phototropic-responsive NPH3 family protein] |
0.338 |
0.005 |
| 88 |
247905_at |
AT5G57400
|
unknown |
0.337 |
-0.028 |
| 89 |
257798_at |
AT3G15950
|
NAI2 |
0.337 |
0.266 |
| 90 |
263236_at |
AT1G10470
|
ARR4, response regulator 4, ATRR1, RESPONCE REGULATOR 1, IBC7, INDUCED BY CYTOKININ 7, MEE7, maternal effect embryo arrest 7 |
0.336 |
-0.054 |
| 91 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.334 |
0.032 |
| 92 |
263664_at |
AT1G04250
|
AXR3, AUXIN RESISTANT 3, IAA17, indole-3-acetic acid inducible 17 |
0.331 |
-0.100 |
| 93 |
255818_at |
AT2G33570
|
GALS1, galactan synthase 1 |
0.325 |
0.026 |
| 94 |
266209_at |
AT2G27550
|
ATC, centroradialis |
-0.905 |
-0.031 |
| 95 |
256349_at |
AT1G54890
|
[Late embryogenesis abundant (LEA) protein-related] |
-0.775 |
-0.019 |
| 96 |
265769_at |
AT2G48090
|
unknown |
-0.667 |
-0.055 |
| 97 |
247634_at |
AT5G60520
|
[Late embryogenesis abundant (LEA) protein-related] |
-0.651 |
-0.022 |
| 98 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
-0.631 |
-0.140 |
| 99 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
-0.602 |
0.030 |
| 100 |
245385_at |
AT4G14020
|
[Rapid alkalinization factor (RALF) family protein] |
-0.589 |
-0.066 |
| 101 |
250469_at |
AT5G10130
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.542 |
-0.039 |
| 102 |
247922_at |
AT5G57500
|
[Galactosyltransferase family protein] |
-0.531 |
0.049 |
| 103 |
251298_at |
AT3G62040
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.531 |
-0.056 |
| 104 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
-0.529 |
0.038 |
| 105 |
263481_at |
AT2G04025
|
GLV7, GOLVEN 7, RGF3, root meristem growth factor 3 |
-0.519 |
0.010 |
| 106 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-0.510 |
-0.040 |
| 107 |
257375_at |
AT2G38640
|
unknown |
-0.504 |
-0.122 |
| 108 |
267457_at |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
-0.499 |
-0.001 |
| 109 |
253483_at |
AT4G31910
|
BAT1, BR-related AcylTransferase1, PIZ, PIZZA |
-0.487 |
0.008 |
| 110 |
262728_at |
AT1G75820
|
ATCLV1, CLV1, CLAVATA 1, FAS3, FASCIATA 3, FLO5, FLOWER DEVELOPMENT 5 |
-0.484 |
-0.031 |
| 111 |
258362_at |
AT3G14280
|
unknown |
-0.469 |
0.054 |
| 112 |
246125_at |
AT5G19875
|
unknown |
-0.454 |
-0.045 |
| 113 |
259275_at |
AT3G01060
|
unknown |
-0.454 |
-0.094 |
| 114 |
265856_at |
AT2G42430
|
ASL18, ASYMMETRIC LEAVES2-LIKE 18, LBD16, lateral organ boundaries-domain 16 |
-0.429 |
-0.048 |
| 115 |
258293_at |
AT3G23430
|
ATPHO1, ARABIDOPSIS PHOSPHATE 1, PHO1, phosphate 1 |
-0.427 |
-0.032 |
| 116 |
245891_at |
AT5G09220
|
AAP2, amino acid permease 2 |
-0.409 |
0.004 |
| 117 |
250002_at |
AT5G18690
|
AGP25, arabinogalactan protein 25, ATAGP25, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEINS 25 |
-0.396 |
0.103 |
| 118 |
262154_at |
AT1G52700
|
[alpha/beta-Hydrolases superfamily protein] |
-0.390 |
0.074 |
| 119 |
249126_at |
AT5G43380
|
TOPP6, type one serine/threonine protein phosphatase 6 |
-0.390 |
-0.036 |
| 120 |
250438_at |
AT5G10580
|
unknown |
-0.385 |
-0.055 |
| 121 |
249440_at |
AT5G40030
|
[Protein kinase superfamily protein] |
-0.385 |
-0.048 |
| 122 |
260077_at |
AT1G73620
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.383 |
0.177 |
| 123 |
253629_at |
AT4G30450
|
[glycine-rich protein] |
-0.381 |
-0.070 |
| 124 |
245488_at |
AT4G16270
|
[Peroxidase superfamily protein] |
-0.380 |
-0.036 |
| 125 |
264323_at |
AT1G04180
|
YUC9, YUCCA 9 |
-0.365 |
-0.009 |
| 126 |
261327_at |
AT1G44830
|
[Integrase-type DNA-binding superfamily protein] |
-0.364 |
0.046 |
| 127 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
-0.362 |
0.008 |
| 128 |
250231_at |
AT5G13910
|
LEP, LEAFY PETIOLE |
-0.359 |
-0.008 |
| 129 |
260076_at |
AT1G73630
|
[EF hand calcium-binding protein family] |
-0.355 |
-0.134 |
| 130 |
263482_at |
AT2G03980
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.353 |
-0.027 |
| 131 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
-0.352 |
0.610 |
| 132 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
-0.345 |
0.053 |
| 133 |
252988_at |
AT4G38410
|
[Dehydrin family protein] |
-0.342 |
-0.050 |
| 134 |
263465_at |
AT2G31940
|
unknown |
-0.341 |
-0.061 |
| 135 |
264889_at |
AT1G23050
|
hydroxyproline-rich glycoprotein family protein |
-0.335 |
-0.051 |
| 136 |
246390_at |
AT1G77330
|
ACO5, ACC oxidase 5 |
-0.332 |
-0.068 |
| 137 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
-0.332 |
0.021 |
| 138 |
266301_at |
AT2G26960
|
AtMYB81, myb domain protein 81, MYB81, myb domain protein 81 |
-0.318 |
-0.016 |
| 139 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
-0.316 |
0.350 |
| 140 |
249059_at |
AT5G44530
|
[Subtilase family protein] |
-0.312 |
-0.161 |
| 141 |
261203_at |
AT1G12845
|
unknown |
-0.311 |
0.101 |
| 142 |
255645_at |
AT4G00880
|
SAUR31, SMALL AUXIN UPREGULATED RNA 31 |
-0.305 |
-0.103 |
| 143 |
247553_at |
AT5G60910
|
AGL8, AGAMOUS-like 8, FUL, FRUITFULL |
-0.305 |
-0.071 |
| 144 |
248961_at |
AT5G45650
|
[subtilase family protein] |
-0.301 |
-0.035 |
| 145 |
254037_at |
AT4G25760
|
ATGDU2, glutamine dumper 2, GDU2, glutamine dumper 2 |
-0.300 |
0.025 |
| 146 |
248807_at |
AT5G47500
|
PME5, pectin methylesterase 5 |
-0.299 |
0.268 |
| 147 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
-0.299 |
-0.076 |
| 148 |
251919_at |
AT3G53800
|
Fes1B, Fes1B |
-0.299 |
0.104 |
| 149 |
256100_at |
AT1G13750
|
[Purple acid phosphatases superfamily protein] |
-0.294 |
0.001 |
| 150 |
248370_at |
AT5G52170
|
HDG7, homeodomain GLABROUS 7 |
-0.291 |
-0.012 |
| 151 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
-0.290 |
0.095 |
| 152 |
253515_at |
AT4G31320
|
SAUR37, SMALL AUXIN UPREGULATED RNA 37 |
-0.289 |
-0.036 |
| 153 |
256574_at |
AT3G14780
|
unknown |
-0.287 |
-0.055 |
| 154 |
266561_at |
AT2G23960
|
[Class I glutamine amidotransferase-like superfamily protein] |
-0.287 |
0.013 |
| 155 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.284 |
-0.043 |
| 156 |
264626_at |
AT1G65620
|
AS2, ASYMMETRIC LEAVES 2 |
-0.284 |
-0.006 |
| 157 |
262969_at |
AT1G75710
|
[C2H2-like zinc finger protein] |
-0.283 |
-0.051 |
| 158 |
253837_at |
AT4G27850
|
[Glycine-rich protein family] |
-0.283 |
-0.060 |
| 159 |
258332_at |
AT3G16180
|
NRT1.12, nitrate transporter 1.12 |
-0.278 |
0.102 |
| 160 |
259163_at |
AT3G01490
|
[Protein kinase superfamily protein] |
-0.277 |
0.029 |
| 161 |
251317_at |
AT3G61490
|
[Pectin lyase-like superfamily protein] |
-0.276 |
0.054 |
| 162 |
250493_at |
AT5G09800
|
[ARM repeat superfamily protein] |
-0.273 |
-0.001 |
| 163 |
262752_at |
AT1G16330
|
CYCB3;1, cyclin b3;1 |
-0.272 |
0.043 |
| 164 |
245399_at |
AT4G17340
|
DELTA-TIP2, TIP2;2, tonoplast intrinsic protein 2;2 |
-0.269 |
-0.069 |
| 165 |
245980_at |
AT5G13140
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.267 |
0.029 |
| 166 |
252882_at |
AT4G39675
|
unknown |
-0.265 |
-0.064 |
| 167 |
260558_at |
AT2G43600
|
[Chitinase family protein] |
-0.259 |
-0.021 |
| 168 |
266736_at |
AT2G46960
|
CYP709B1, cytochrome P450, family 709, subfamily B, polypeptide 1 |
-0.258 |
-0.012 |
| 169 |
250764_at |
AT5G05960
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.255 |
0.106 |
| 170 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.252 |
-0.036 |
| 171 |
249190_at |
AT5G42750
|
BKI1, BRI1 kinase inhibitor 1 |
-0.250 |
-0.054 |
| 172 |
258596_at |
AT3G04510
|
LSH2, LIGHT SENSITIVE HYPOCOTYLS 2 |
-0.249 |
-0.025 |
| 173 |
258611_at |
AT3G02860
|
[zinc ion binding] |
-0.248 |
-0.096 |
| 174 |
258560_at |
AT3G06020
|
FAF4, FANTASTIC FOUR 4 |
-0.245 |
0.056 |
| 175 |
255578_at |
AT4G01450
|
UMAMIT30, Usually multiple acids move in and out Transporters 30 |
-0.243 |
-0.070 |
| 176 |
246429_at |
AT5G17450
|
HIPP21, heavy metal associated isoprenylated plant protein 21 |
-0.242 |
0.001 |
| 177 |
264313_at |
AT1G70410
|
ATBCA4, BETA CARBONIC ANHYDRASE 4, BCA4, beta carbonic anhydrase 4, CA4, BETA CARBONIC ANHYDRASE 4 |
-0.240 |
-0.058 |
| 178 |
263452_at |
AT2G22190
|
TPPE, trehalose-6-phosphate phosphatase E |
-0.238 |
-0.123 |
| 179 |
257026_at |
AT3G19200
|
unknown |
-0.237 |
0.047 |
| 180 |
260434_at |
AT1G68330
|
unknown |
-0.235 |
0.108 |
| 181 |
259171_at |
AT3G03590
|
[SWIB/MDM2 domain superfamily protein] |
-0.234 |
0.010 |
| 182 |
266280_at |
AT2G29260
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.231 |
-0.039 |
| 183 |
251592_at |
AT3G57670
|
NTT, NO TRANSMITTING TRACT, WIP2, WIP domain protein 2 |
-0.231 |
-0.069 |
| 184 |
247038_at |
AT5G67160
|
EPS1, ENHANCED PSEUDOMONAS SUSCEPTIBILTY 1 |
-0.230 |
-0.046 |
| 185 |
263610_at |
AT2G16230
|
[O-Glycosyl hydrolases family 17 protein] |
-0.230 |
-0.041 |