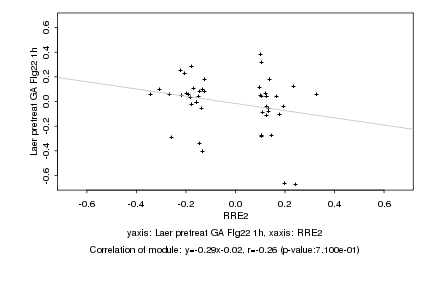
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
RRE2 |
Log2 signal ratio
Laer pretreat GA Flg22 1h |
| 1 |
257592_at |
AT3G24982
|
ATRLP40, receptor like protein 40, RLP40, receptor like protein 40 |
0.326 |
0.061 |
| 2 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
0.241 |
-0.665 |
| 3 |
257163_at |
AT3G24310
|
ATMYB71, MYB DOMAIN PROTEIN 71, MYB305, myb domain protein 305 |
0.234 |
0.128 |
| 4 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
0.198 |
-0.664 |
| 5 |
251647_at |
AT3G57770
|
[Protein kinase superfamily protein] |
0.193 |
-0.034 |
| 6 |
260385_at |
AT1G74090
|
ATSOT18, DESULFO-GLUCOSINOLATE SULFOTRANSFERASE 18, ATST5B, ARABIDOPSIS SULFOTRANSFERASE 5B, SOT18, desulfo-glucosinolate sulfotransferase 18 |
0.176 |
-0.103 |
| 7 |
249849_at |
AT5G23230
|
NIC2, nicotinamidase 2 |
0.164 |
0.047 |
| 8 |
262940_at |
AT1G79520
|
[Cation efflux family protein] |
0.143 |
-0.271 |
| 9 |
259143_at |
AT3G10190
|
[Calcium-binding EF-hand family protein] |
0.135 |
0.181 |
| 10 |
257672_at |
AT3G20300
|
unknown |
0.133 |
-0.080 |
| 11 |
252740_at |
AT3G43270
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.132 |
-0.054 |
| 12 |
259854_at |
AT1G72200
|
[RING/U-box superfamily protein] |
0.124 |
0.045 |
| 13 |
255237_at |
AT4G05550
|
[Mutator-like transposase family, has a 4.9e-80 P-value blast match to Q9S9W4 /247-408 Pfam PF03108 MuDR family transposase (MuDr-element domain)] |
0.124 |
0.043 |
| 14 |
247946_at |
AT5G57180
|
CIA2, chloroplast import apparatus 2 |
0.124 |
-0.113 |
| 15 |
257369_at |
AT2G35550
|
ATBPC7, BASIC PENTACYSTEINE 7, BBR, BPC7, basic pentacysteine 7 |
0.122 |
-0.034 |
| 16 |
247467_at |
AT5G62130
|
[Per1-like family protein] |
0.121 |
0.068 |
| 17 |
258209_at |
AT3G14060
|
unknown |
0.107 |
-0.088 |
| 18 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
0.105 |
-0.281 |
| 19 |
260634_at |
AT1G62410
|
[MIF4G domain-containing protein] |
0.104 |
0.048 |
| 20 |
266291_at |
AT2G29320
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.103 |
-0.275 |
| 21 |
254664_at |
AT4G18300
|
[Trimeric LpxA-like enzyme] |
0.103 |
0.322 |
| 22 |
256693_at |
AT3G32060
|
[Mutator-like transposase family, has a 4.0e-63 P-value blast match to O80466 /172-336 Pfam PF03108 MuDR family transposase (MuDr-element domain)] |
0.101 |
0.050 |
| 23 |
259596_at |
AT1G28130
|
GH3.17 |
0.099 |
0.387 |
| 24 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.096 |
0.116 |
| 25 |
266383_at |
AT2G14580
|
ATPRB1, basic pathogenesis-related protein 1, PRB1, basic pathogenesis-related protein 1 |
-0.346 |
0.057 |
| 26 |
255937_at |
AT1G12610
|
DDF1, DWARF AND DELAYED FLOWERING 1 |
-0.308 |
0.098 |
| 27 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.267 |
0.060 |
| 28 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
-0.262 |
-0.291 |
| 29 |
249364_at |
AT5G40590
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.222 |
0.256 |
| 30 |
248400_at |
AT5G52020
|
[Integrase-type DNA-binding superfamily protein] |
-0.221 |
0.049 |
| 31 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
-0.209 |
0.234 |
| 32 |
257701_at |
AT3G12710
|
[DNA glycosylase superfamily protein] |
-0.198 |
0.068 |
| 33 |
261101_at |
AT1G63030
|
ddf2, DWARF AND DELAYED FLOWERING 2 |
-0.192 |
0.058 |
| 34 |
264697_at |
AT1G70210
|
ATCYCD1;1, CYCD1;1, CYCLIN D1;1 |
-0.185 |
0.035 |
| 35 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.181 |
-0.022 |
| 36 |
259552_at |
AT1G21320
|
[nucleotide binding] |
-0.180 |
0.291 |
| 37 |
253155_at |
AT4G35720
|
unknown |
-0.171 |
0.106 |
| 38 |
254609_at |
AT4G18970
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.159 |
-0.003 |
| 39 |
246562_at |
AT5G15580
|
LNG1, LONGIFOLIA1, TRM2, TON1 Recruiting Motif 2 |
-0.159 |
-0.007 |
| 40 |
250216_at |
AT5G14090
|
AtLAZY1, LAZY1, LAZY 1 |
-0.152 |
0.048 |
| 41 |
266215_at |
AT2G06850
|
EXGT-A1, endoxyloglucan transferase A1, EXT, ENDOXYLOGLUCAN TRANSFERASE, XTH4, xyloglucan endotransglucosylase/hydrolase 4 |
-0.146 |
-0.333 |
| 42 |
252517_at |
AT3G46340
|
[Leucine-rich repeat protein kinase family protein] |
-0.146 |
0.084 |
| 43 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.139 |
-0.055 |
| 44 |
255096_at |
AT4G08600
|
unknown |
-0.135 |
0.098 |
| 45 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
-0.134 |
-0.403 |
| 46 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
-0.127 |
0.179 |
| 47 |
262543_at |
AT1G34245
|
EPF2, EPIDERMAL PATTERNING FACTOR 2 |
-0.127 |
0.087 |