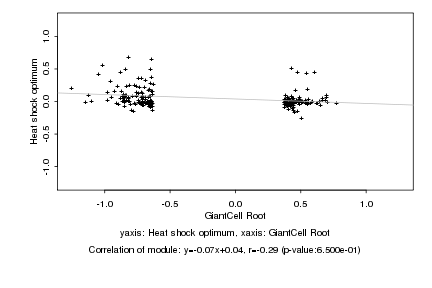
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
GiantCell Root |
Log2 signal ratio
Heat shock optimum |
| 1 |
254130_at |
AT4G24540
|
AGL24, AGAMOUS-like 24 |
0.766 |
-0.026 |
| 2 |
257118_at |
AT3G20180
|
[Copper transport protein family] |
0.700 |
-0.013 |
| 3 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
0.691 |
0.062 |
| 4 |
258981_at |
AT3G08880
|
unknown |
0.690 |
0.096 |
| 5 |
264188_at |
AT1G54690
|
G-H2AX, GAMMA H2AX, GAMMA-H2AX, gamma histone variant H2AX, H2AXB, HTA3, histone H2A 3 |
0.684 |
0.017 |
| 6 |
249147_at |
AT5G43330
|
c-NAD-MDH2, cytosolic-NAD-dependent malate dehydrogenase 2 |
0.663 |
0.030 |
| 7 |
249028_at |
AT5G44740
|
POLH, Y-family DNA polymerase H |
0.661 |
0.055 |
| 8 |
261222_at |
AT1G20120
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.647 |
-0.048 |
| 9 |
250816_at |
AT5G05010
|
[clathrin adaptor complexes medium subunit family protein] |
0.640 |
0.004 |
| 10 |
254403_at |
AT4G21323
|
[Subtilase family protein] |
0.627 |
-0.030 |
| 11 |
247228_at |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
0.614 |
-0.030 |
| 12 |
264019_at |
AT2G21130
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.599 |
0.448 |
| 13 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
0.585 |
0.008 |
| 14 |
249694_at |
AT5G35790
|
G6PD1, glucose-6-phosphate dehydrogenase 1 |
0.578 |
-0.015 |
| 15 |
248801_at |
AT5G47370
|
HAT2 |
0.571 |
-0.024 |
| 16 |
265386_at |
AT2G20930
|
[SNARE-like superfamily protein] |
0.554 |
0.035 |
| 17 |
247460_at |
AT5G62050
|
ATOXA1, ARABIDOPSIS THALIANA HOMOLOG OF YEAST OXIDASE ASSEMBLY 1 (OXA1), OXA1, homolog of yeast oxidase assembly 1 (OXA1), OXA1AT, HOMOLOG OF YEAST OXIDASE ASSEMBLY 1 (OXA1) IN ARABIDOPSIS THALIANA |
0.554 |
-0.023 |
| 18 |
255439_at |
AT4G03080
|
BSL1, BRI1 suppressor 1 (BSU1)-like 1 |
0.550 |
-0.029 |
| 19 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
0.548 |
0.199 |
| 20 |
254245_at |
AT4G23240
|
CRK16, cysteine-rich RLK (RECEPTOR-like protein kinase) 16 |
0.547 |
-0.040 |
| 21 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.543 |
0.433 |
| 22 |
250100_at |
AT5G16570
|
GLN1;4, glutamine synthetase 1;4 |
0.541 |
-0.026 |
| 23 |
260701_at |
AT1G32330
|
ATHSFA1D, heat shock transcription factor A1D, HSFA1D, heat shock transcription factor A1D |
0.535 |
0.029 |
| 24 |
253173_at |
AT4G35110
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.530 |
-0.016 |
| 25 |
263916_at |
AT2G36440
|
unknown |
0.511 |
-0.012 |
| 26 |
260323_at |
AT1G63780
|
IMP4 |
0.507 |
0.008 |
| 27 |
267404_at |
AT2G33980
|
atnudt22, nudix hydrolase homolog 22, NUDT22, nudix hydrolase homolog 22 |
0.506 |
-0.006 |
| 28 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
0.505 |
-0.251 |
| 29 |
262555_at |
AT1G31400
|
[TRAF-like family protein] |
0.504 |
-0.005 |
| 30 |
248305_at |
AT5G52560
|
ATUSP, UDP-sugar pyrophosphorylase, USP, UDP-sugar pyrophosphorylase |
0.491 |
0.017 |
| 31 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
0.490 |
-0.033 |
| 32 |
249064_at |
AT5G44250
|
unknown |
0.488 |
-0.039 |
| 33 |
252086_at |
AT3G52030
|
[F-box family protein with WD40/YVTN repeat doamin] |
0.487 |
0.004 |
| 34 |
254342_at |
AT4G22140
|
EBS, EARLY BOLTING IN SHORT DAYS |
0.486 |
0.038 |
| 35 |
254415_at |
AT4G21326
|
ATSBT3.12, subtilase 3.12, SBT3.12, subtilase 3.12 |
0.486 |
0.041 |
| 36 |
247691_at |
AT5G59720
|
HSP18.2, heat shock protein 18.2 |
0.486 |
0.068 |
| 37 |
255720_at |
AT1G32060
|
PRK, phosphoribulokinase |
0.486 |
-0.036 |
| 38 |
250940_at |
AT5G03310
|
SAUR44, SMALL AUXIN UPREGULATED RNA 44 |
0.485 |
-0.014 |
| 39 |
247563_at |
AT5G61130
|
AtPDCB1, PDCB1, plasmodesmata callose-binding protein 1 |
0.478 |
-0.015 |
| 40 |
256976_at |
AT3G21020
|
[copia-like retrotransposon family, has a 5.4e-14 P-value blast match to gb|AAO73529.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
0.478 |
0.027 |
| 41 |
251558_at |
AT3G57810
|
[Cysteine proteinases superfamily protein] |
0.475 |
0.022 |
| 42 |
257801_at |
AT3G18750
|
ATWNK6, ARABIDOPSIS THALIANA WITH NO K 6, WNK6, with no lysine (K) kinase 6, ZIK5 |
0.472 |
-0.150 |
| 43 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
0.470 |
0.454 |
| 44 |
247331_at |
AT5G63530
|
ATFP3, ARABIDOPSIS THALIANA FARNESYLATED PROTEIN 3, FP3, farnesylated protein 3 |
0.467 |
-0.013 |
| 45 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
0.466 |
0.029 |
| 46 |
262528_at |
AT1G17260
|
AHA10, autoinhibited H(+)-ATPase isoform 10 |
0.461 |
0.016 |
| 47 |
261014_at |
AT1G26460
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.461 |
0.003 |
| 48 |
262948_at |
AT1G75560
|
[zinc knuckle (CCHC-type) family protein] |
0.459 |
-0.011 |
| 49 |
258406_at |
AT3G17611
|
ATRBL10, RHOMBOID-like protein 10, ATRBL14, RHOMBOID-like protein 14, RBL10, RHOMBOID-like protein 10, RBL14, RHOMBOID-like protein 14 |
0.457 |
0.180 |
| 50 |
245152_at |
AT2G47490
|
ATNDT1, ARABIDOPSIS THALIANA NAD+ TRANSPORTER 1, NDT1, NAD+ transporter 1 |
0.448 |
-0.160 |
| 51 |
259212_at |
AT3G09180
|
unknown |
0.446 |
0.004 |
| 52 |
259913_at |
AT1G72660
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.443 |
0.023 |
| 53 |
251915_at |
AT3G53940
|
[Mitochondrial substrate carrier family protein] |
0.442 |
0.056 |
| 54 |
266627_at |
AT2G35340
|
MEE29, maternal effect embryo arrest 29 |
0.442 |
-0.005 |
| 55 |
266076_at |
AT2G40700
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.440 |
-0.012 |
| 56 |
245528_at |
AT4G15530
|
PPDK, pyruvate orthophosphate dikinase |
0.440 |
-0.132 |
| 57 |
258338_at |
AT3G16150
|
ASPGB1, asparaginase B1 |
0.439 |
-0.052 |
| 58 |
266689_at |
AT2G19930
|
[RNA-dependent RNA polymerase family protein] |
0.439 |
0.008 |
| 59 |
261377_at |
AT1G18850
|
unknown |
0.437 |
-0.017 |
| 60 |
251091_at |
AT5G01410
|
ATPDX1, ATPDX1.3, ARABIDOPSIS THALIANA PYRIDOXINE BIOSYNTHESIS 1.3, PDX1, PDX1.3, PYRIDOXINE BIOSYNTHESIS 1.3, RSR4, REDUCED SUGAR RESPONSE 4 |
0.437 |
-0.078 |
| 61 |
246151_at |
AT5G19950
|
unknown |
0.433 |
0.059 |
| 62 |
251365_at |
AT3G61310
|
AHL11, AT-hook motif nuclear localized protein 11 |
0.432 |
-0.025 |
| 63 |
254134_at |
AT4G24830
|
[arginosuccinate synthase family] |
0.432 |
0.001 |
| 64 |
245110_at |
AT2G41550
|
[Rho termination factor] |
0.431 |
0.026 |
| 65 |
263406_at |
AT2G04160
|
AIR3, AUXIN-INDUCED IN ROOT CULTURES 3 |
0.431 |
-0.096 |
| 66 |
245990_at |
AT5G20640
|
unknown |
0.429 |
-0.032 |
| 67 |
253488_at |
AT4G31610
|
ATREM1, REPRODUCTIVE MERISTEM 1, REM1, REPRODUCTIVE MERISTEM 1 |
0.428 |
-0.011 |
| 68 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
0.428 |
-0.049 |
| 69 |
248086_at |
AT5G55490
|
ATGEX1, GAMETE EXPRESSED PROTEIN 1, GEX1, gamete expressed protein 1 |
0.426 |
-0.040 |
| 70 |
259037_at |
AT3G09350
|
Fes1A, Fes1A |
0.425 |
0.509 |
| 71 |
247709_at |
AT5G59250
|
[Major facilitator superfamily protein] |
0.425 |
-0.021 |
| 72 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.425 |
0.092 |
| 73 |
259207_at |
AT3G09050
|
unknown |
0.422 |
-0.039 |
| 74 |
261969_at |
AT1G65950
|
[Protein kinase superfamily protein] |
0.421 |
-0.069 |
| 75 |
251265_at |
AT3G62310
|
[RNA helicase family protein] |
0.420 |
-0.054 |
| 76 |
259907_at |
AT1G60860
|
AGD2, ARF-GAP domain 2 |
0.420 |
0.024 |
| 77 |
250290_at |
AT5G13310
|
unknown |
0.419 |
0.023 |
| 78 |
256805_at |
AT3G20930
|
ORRM1, Organelle RRM protein 1 |
0.417 |
-0.031 |
| 79 |
251271_at |
AT3G62050
|
[Putative endonuclease or glycosyl hydrolase] |
0.416 |
0.024 |
| 80 |
249791_at |
AT5G23810
|
AAP7, amino acid permease 7 |
0.413 |
0.033 |
| 81 |
259088_at |
AT3G04970
|
[DHHC-type zinc finger family protein] |
0.411 |
-0.028 |
| 82 |
259469_at |
AT1G19100
|
AtMORC6, DMS11, DEFECTIVE IN MERISTEM SILENCING 11, MORC6, Microrchidia 6 |
0.411 |
-0.016 |
| 83 |
253932_at |
AT4G26800
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.406 |
-0.015 |
| 84 |
260771_at |
AT1G49160
|
WNK7 |
0.405 |
-0.118 |
| 85 |
266716_at |
AT2G46820
|
CURT1B, CURVATURE THYLAKOID 1B, PSAP, PSI-P, photosystem I P subunit, PTAC8, PLASTID TRANSCRIPTIONALLY ACTIVE 8, TMP14, THYLAKOID MEMBRANE PHOSPHOPROTEIN OF 14 KDA |
0.402 |
-0.019 |
| 86 |
261263_at |
AT1G26790
|
[Dof-type zinc finger DNA-binding family protein] |
0.400 |
-0.030 |
| 87 |
253116_at |
AT4G35850
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.399 |
0.048 |
| 88 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
0.399 |
-0.039 |
| 89 |
262584_at |
AT1G15440
|
ATPWP2, PERIODIC TRYPTOPHAN PROTEIN 2, PWP2, periodic tryptophan protein 2 |
0.398 |
0.073 |
| 90 |
264742_at |
AT1G62130
|
[AAA-type ATPase family protein] |
0.398 |
-0.067 |
| 91 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.397 |
0.026 |
| 92 |
265263_at |
AT2G42940
|
AHL16, AT-hook motif nuclear-localized protein 16, TEK, TRANSPOSABLE ELEMENT SILENCING VIA AT-HOOK |
0.396 |
-0.051 |
| 93 |
246145_at |
AT5G19880
|
[Peroxidase superfamily protein] |
0.394 |
-0.045 |
| 94 |
266137_at |
AT2G45010
|
[PLAC8 family protein] |
0.392 |
0.018 |
| 95 |
251099_at |
AT5G01660
|
unknown |
0.391 |
-0.019 |
| 96 |
247038_at |
AT5G67160
|
EPS1, ENHANCED PSEUDOMONAS SUSCEPTIBILTY 1 |
0.388 |
-0.046 |
| 97 |
260916_at |
AT1G02475
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.386 |
-0.048 |
| 98 |
252941_at |
AT4G39280
|
[phenylalanyl-tRNA synthetase, putative / phenylalanine--tRNA ligase, putative] |
0.385 |
-0.019 |
| 99 |
251346_at |
AT3G60980
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.385 |
-0.041 |
| 100 |
261417_at |
AT1G07700
|
[Thioredoxin superfamily protein] |
0.384 |
-0.040 |
| 101 |
254925_at |
AT4G11380
|
[Adaptin family protein] |
0.383 |
-0.001 |
| 102 |
264731_at |
AT1G62150
|
[Mitochondrial transcription termination factor family protein] |
0.383 |
-0.040 |
| 103 |
248173_at |
AT5G54580
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.383 |
-0.027 |
| 104 |
248556_at |
AT5G50350
|
unknown |
0.382 |
0.005 |
| 105 |
246933_at |
AT5G25160
|
ZFP3, zinc finger protein 3 |
0.382 |
0.028 |
| 106 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
0.381 |
-0.032 |
| 107 |
258160_at |
AT3G17820
|
ATGSKB6, ARABIDOPSIS THALIANA GLUTAMINE SYNTHASE CLONE KB6, GLN1.3, glutamine synthetase 1.3, GLN1;3, GLUTAMINE SYNTHETASE 1;3 |
0.380 |
0.096 |
| 108 |
253084_at |
AT4G36260
|
SRS2, SHI RELATED SEQUENCE 2, STY2, STYLISH 2 |
0.379 |
0.001 |
| 109 |
265908_at |
AT4G00270
|
[DNA-binding storekeeper protein-related transcriptional regulator] |
0.377 |
0.035 |
| 110 |
258965_at |
AT3G10530
|
[Transducin/WD40 repeat-like superfamily protein] |
0.375 |
0.041 |
| 111 |
264298_at |
AT1G78690
|
At1g78690p |
0.375 |
-0.033 |
| 112 |
266604_at |
AT2G46030
|
UBC6, ubiquitin-conjugating enzyme 6 |
0.374 |
-0.008 |
| 113 |
258673_at |
AT3G08620
|
[RNA-binding KH domain-containing protein] |
0.372 |
-0.088 |
| 114 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.372 |
-0.007 |
| 115 |
253832_at |
AT4G27654
|
unknown |
-1.260 |
0.211 |
| 116 |
259264_at |
AT3G01260
|
[Galactose mutarotase-like superfamily protein] |
-1.154 |
-0.001 |
| 117 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
-1.130 |
0.104 |
| 118 |
245891_at |
AT5G09220
|
AAP2, amino acid permease 2 |
-1.103 |
0.004 |
| 119 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
-1.054 |
0.418 |
| 120 |
253259_at |
AT4G34410
|
RRTF1, redox responsive transcription factor 1 |
-1.021 |
0.557 |
| 121 |
261476_at |
AT1G14480
|
[Ankyrin repeat family protein] |
-0.985 |
0.141 |
| 122 |
254896_at |
AT4G11880
|
AGL14, AGAMOUS-like 14, XAL2, XAANTAL2 |
-0.984 |
0.021 |
| 123 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
-0.957 |
0.308 |
| 124 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
-0.956 |
0.068 |
| 125 |
267393_at |
AT2G44500
|
[O-fucosyltransferase family protein] |
-0.927 |
0.169 |
| 126 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
-0.913 |
-0.020 |
| 127 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
-0.909 |
0.232 |
| 128 |
247755_at |
AT5G59090
|
ATSBT4.12, subtilase 4.12, SBT4.12, subtilase 4.12 |
-0.898 |
-0.039 |
| 129 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
-0.884 |
0.051 |
| 130 |
257644_at |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
-0.880 |
0.457 |
| 131 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
-0.878 |
0.160 |
| 132 |
254158_at |
AT4G24380
|
[INVOLVED IN: 10-formyltetrahydrofolate biosynthetic process, folic acid and derivative biosynthetic process] |
-0.867 |
0.092 |
| 133 |
252193_at |
AT3G50060
|
MYB77, myb domain protein 77 |
-0.866 |
0.015 |
| 134 |
256442_at |
AT3G10930
|
unknown |
-0.863 |
0.050 |
| 135 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
-0.860 |
0.122 |
| 136 |
250869_at |
AT5G03840
|
TFL-1, TERMINAL FLOWER 1, TFL1, TERMINAL FLOWER 1 |
-0.856 |
0.014 |
| 137 |
250956_at |
AT5G03210
|
AtDIP2, DIP2, DBP-interacting protein 2 |
-0.856 |
-0.075 |
| 138 |
246984_at |
AT5G67310
|
CYP81G1, cytochrome P450, family 81, subfamily G, polypeptide 1 |
-0.856 |
0.031 |
| 139 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
-0.853 |
-0.013 |
| 140 |
250098_at |
AT5G17350
|
unknown |
-0.851 |
0.068 |
| 141 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
-0.845 |
0.107 |
| 142 |
260744_at |
AT1G15010
|
unknown |
-0.844 |
0.501 |
| 143 |
265680_at |
AT2G32150
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.841 |
0.093 |
| 144 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
-0.841 |
0.027 |
| 145 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
-0.839 |
0.237 |
| 146 |
253191_at |
AT4G35350
|
XCP1, xylem cysteine peptidase 1 |
-0.825 |
0.066 |
| 147 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
-0.822 |
0.680 |
| 148 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
-0.821 |
0.006 |
| 149 |
256021_at |
AT1G58270
|
ZW9 |
-0.820 |
0.057 |
| 150 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
-0.818 |
0.249 |
| 151 |
245041_at |
AT2G26530
|
AR781 |
-0.811 |
0.012 |
| 152 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
-0.809 |
-0.031 |
| 153 |
255129_at |
AT4G08290
|
UMAMIT20, Usually multiple acids move in and out Transporters 20 |
-0.802 |
-0.137 |
| 154 |
245906_at |
AT5G11070
|
unknown |
-0.791 |
0.079 |
| 155 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
-0.785 |
-0.140 |
| 156 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
-0.775 |
0.258 |
| 157 |
245550_at |
AT4G15330
|
CYP705A1, cytochrome P450, family 705, subfamily A, polypeptide 1 |
-0.774 |
-0.028 |
| 158 |
257458_at |
AT2G05400
|
[Ubiquitin-specific protease family C19-related protein] |
-0.765 |
-0.038 |
| 159 |
245252_at |
AT4G17500
|
ATERF-1, ethylene responsive element binding factor 1, ERF-1, ethylene responsive element binding factor 1 |
-0.762 |
0.089 |
| 160 |
264617_at |
AT2G17660
|
[RPM1-interacting protein 4 (RIN4) family protein] |
-0.761 |
0.102 |
| 161 |
265723_at |
AT2G32140
|
[transmembrane receptors] |
-0.761 |
0.245 |
| 162 |
259518_at |
AT1G20510
|
OPCL1, OPC-8:0 CoA ligase1 |
-0.758 |
0.144 |
| 163 |
263867_at |
AT2G36830
|
GAMMA-TIP, gamma tonoplast intrinsic protein, GAMMA-TIP1, GAMMA TONOPLAST INTRINSIC PROTEIN 1, TIP1;1, TONOPLAST INTRINSIC PROTEIN 1;1 |
-0.748 |
0.019 |
| 164 |
262211_at |
AT1G74930
|
ORA47 |
-0.748 |
0.138 |
| 165 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
-0.747 |
0.356 |
| 166 |
267381_at |
AT2G26190
|
[calmodulin-binding family protein] |
-0.745 |
-0.008 |
| 167 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
-0.741 |
0.222 |
| 168 |
259774_at |
AT1G29520
|
[AWPM-19-like family protein] |
-0.739 |
-0.027 |
| 169 |
267101_at |
AT2G41480
|
AtPRX25, PRX25, peroxidase 25 |
-0.736 |
-0.011 |
| 170 |
248904_at |
AT5G46295
|
unknown |
-0.730 |
-0.017 |
| 171 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
-0.725 |
0.360 |
| 172 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
-0.723 |
0.069 |
| 173 |
251804_at |
AT3G55430
|
[O-Glycosyl hydrolases family 17 protein] |
-0.723 |
0.146 |
| 174 |
247047_at |
AT5G66650
|
unknown |
-0.719 |
-0.016 |
| 175 |
266276_at |
AT2G29330
|
TRI, tropinone reductase |
-0.719 |
-0.030 |
| 176 |
266732_at |
AT2G03240
|
[EXS (ERD1/XPR1/SYG1) family protein] |
-0.717 |
0.009 |
| 177 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
-0.717 |
-0.053 |
| 178 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.716 |
0.129 |
| 179 |
252045_at |
AT3G52450
|
AtPUB22, PUB22, plant U-box 22 |
-0.715 |
0.036 |
| 180 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
-0.713 |
0.080 |
| 181 |
261060_at |
AT1G17340
|
[Phosphoinositide phosphatase family protein] |
-0.709 |
0.071 |
| 182 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-0.708 |
-0.057 |
| 183 |
246270_at |
AT4G36500
|
unknown |
-0.696 |
-0.013 |
| 184 |
245244_at |
AT1G44350
|
ILL6, IAA-leucine resistant (ILR)-like gene 6 |
-0.696 |
0.219 |
| 185 |
261606_at |
AT1G49570
|
[Peroxidase superfamily protein] |
-0.693 |
-0.020 |
| 186 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
-0.692 |
0.075 |
| 187 |
256159_at |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
-0.691 |
0.328 |
| 188 |
247693_at |
AT5G59730
|
ATEXO70H7, exocyst subunit exo70 family protein H7, EXO70H7, exocyst subunit exo70 family protein H7 |
-0.691 |
0.046 |
| 189 |
262813_at |
AT1G11670
|
[MATE efflux family protein] |
-0.685 |
-0.025 |
| 190 |
256093_at |
AT1G20823
|
[RING/U-box superfamily protein] |
-0.677 |
0.011 |
| 191 |
264729_at |
AT1G22990
|
HIPP22, heavy metal associated isoprenylated plant protein 22 |
-0.675 |
-0.016 |
| 192 |
249042_at |
AT5G44350
|
[ethylene-responsive nuclear protein -related] |
-0.672 |
-0.044 |
| 193 |
262756_at |
AT1G16370
|
ATOCT6, ARABIDOPSIS THALIANA ORGANIC CATION/CARNITINE TRANSPORTER 6, OCT6, organic cation/carnitine transporter 6 |
-0.672 |
0.184 |
| 194 |
260429_at |
AT1G72450
|
JAZ6, jasmonate-zim-domain protein 6, TIFY11B, TIFY DOMAIN PROTEIN 11B |
-0.669 |
-0.015 |
| 195 |
253891_at |
AT4G27720
|
[Major facilitator superfamily protein] |
-0.668 |
-0.007 |
| 196 |
251507_at |
AT3G59080
|
[Eukaryotic aspartyl protease family protein] |
-0.667 |
0.075 |
| 197 |
256340_at |
AT1G72070
|
[Chaperone DnaJ-domain superfamily protein] |
-0.667 |
-0.015 |
| 198 |
253301_at |
AT4G33720
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.664 |
-0.073 |
| 199 |
250158_at |
AT5G15190
|
unknown |
-0.664 |
0.088 |
| 200 |
252327_at |
AT3G48740
|
AtSWEET11, SWEET11 |
-0.663 |
-0.068 |
| 201 |
266526_at |
AT2G16980
|
[Major facilitator superfamily protein] |
-0.661 |
-0.015 |
| 202 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
-0.661 |
0.190 |
| 203 |
245399_at |
AT4G17340
|
DELTA-TIP2, TIP2;2, tonoplast intrinsic protein 2;2 |
-0.658 |
-0.069 |
| 204 |
265611_at |
AT2G25510
|
unknown |
-0.657 |
-0.016 |
| 205 |
254644_at |
AT4G18510
|
CLE2, CLAVATA3/ESR-related 2 |
-0.657 |
-0.008 |
| 206 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
-0.656 |
0.285 |
| 207 |
261224_at |
AT1G20160
|
ATSBT5.2 |
-0.656 |
-0.086 |
| 208 |
252906_at |
AT4G39640
|
GGT1, gamma-glutamyl transpeptidase 1 |
-0.655 |
-0.047 |
| 209 |
255937_at |
AT1G12610
|
DDF1, DWARF AND DELAYED FLOWERING 1 |
-0.655 |
0.493 |
| 210 |
245779_at |
AT1G73510
|
unknown |
-0.653 |
0.027 |
| 211 |
246949_at |
AT5G25140
|
CYP71B13, cytochrome P450, family 71, subfamily B, polypeptide 13 |
-0.652 |
-0.015 |
| 212 |
247789_at |
AT5G58680
|
[ARM repeat superfamily protein] |
-0.652 |
0.021 |
| 213 |
264655_at |
AT1G09070
|
(AT)SRC2, SOYBEAN GENE REGULATED BY COLD-2, AtSRC2, SRC2, soybean gene regulated by cold-2 |
-0.650 |
0.091 |
| 214 |
263935_at |
AT2G35930
|
AtPUB23, PUB23, plant U-box 23 |
-0.649 |
0.169 |
| 215 |
258516_at |
AT3G06490
|
AtMYB108, myb domain protein 108, BOS1, BOTRYTIS-SUSCEPTIBLE1, MYB108, myb domain protein 108 |
-0.648 |
0.012 |
| 216 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
-0.647 |
0.371 |
| 217 |
255160_at |
AT4G07820
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.646 |
-0.001 |
| 218 |
266209_at |
AT2G27550
|
ATC, centroradialis |
-0.645 |
-0.031 |
| 219 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
-0.644 |
0.650 |
| 220 |
258414_at |
AT3G17380
|
[TRAF-like family protein] |
-0.643 |
0.012 |
| 221 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
-0.643 |
0.015 |
| 222 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.642 |
-0.068 |
| 223 |
253859_at |
AT4G27657
|
unknown |
-0.640 |
0.122 |
| 224 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
-0.639 |
0.157 |
| 225 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
-0.639 |
-0.123 |
| 226 |
252458_at |
AT3G47210
|
unknown |
-0.637 |
-0.024 |
| 227 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
-0.631 |
0.276 |