|
probeID |
AGICode |
Annotation |
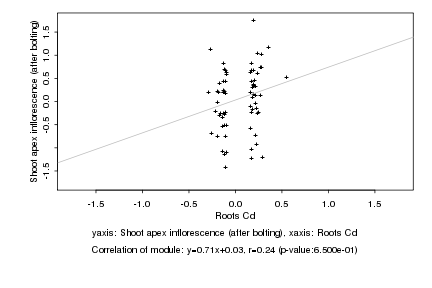
Log2 signal ratio
Roots Cd |
Log2 signal ratio
Shoot apex inflorescence (after bolting) |
| 1 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
0.547 |
0.539 |
| 2 |
261660_at |
AT1G18370
|
ATNACK1, ARABIDOPSIS NPK1-ACTIVATING KINESIN 1, HIK, HINKEL, NACK1, NPK1-ACTIVATING KINESIN 1 |
0.355 |
1.187 |
| 3 |
261432_at |
AT1G07680
|
unknown |
0.287 |
-1.206 |
| 4 |
252692_at |
AT3G43960
|
[Cysteine proteinases superfamily protein] |
0.279 |
0.755 |
| 5 |
257740_at |
AT3G27330
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.275 |
1.031 |
| 6 |
258867_at |
AT3G03130
|
unknown |
0.263 |
0.139 |
| 7 |
252077_at |
AT3G51720
|
unknown |
0.262 |
0.756 |
| 8 |
247942_at |
AT5G57120
|
unknown |
0.245 |
-0.226 |
| 9 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
0.235 |
0.625 |
| 10 |
245568_at |
AT4G14650
|
unknown |
0.234 |
-0.246 |
| 11 |
257813_at |
AT3G25100
|
CDC45, cell division cycle 45 |
0.228 |
1.040 |
| 12 |
252510_at |
AT3G46270
|
[receptor protein kinase-related] |
0.225 |
-0.917 |
| 13 |
259930_at |
AT1G34355
|
ATPS1, PARALLEL SPINDLE 1, PS1, PARALLEL SPINDLE 1 |
0.222 |
-0.147 |
| 14 |
248443_at |
AT5G51310
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.215 |
-0.037 |
| 15 |
249588_at |
AT5G37790
|
[Protein kinase superfamily protein] |
0.212 |
-0.729 |
| 16 |
259686_at |
AT1G63100
|
[GRAS family transcription factor] |
0.209 |
0.332 |
| 17 |
264895_at |
AT1G23100
|
[GroES-like family protein] |
0.206 |
0.133 |
| 18 |
252442_at |
AT3G46940
|
DUT1, DUTP-PYROPHOSPHATASE-LIKE 1 |
0.196 |
0.457 |
| 19 |
256864_at |
AT3G23890
|
ATTOPII, TOPII, topoisomerase II |
0.192 |
0.357 |
| 20 |
253743_at |
AT4G28940
|
[Phosphorylase superfamily protein] |
0.189 |
0.375 |
| 21 |
253156_at |
AT4G35730
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.187 |
0.169 |
| 22 |
265694_at |
AT2G24440
|
[selenium binding] |
0.187 |
0.674 |
| 23 |
255513_at |
AT4G02060
|
MCM7, PRL, PROLIFERA |
0.184 |
1.771 |
| 24 |
265909_at |
AT2G25720
|
unknown |
0.183 |
0.098 |
| 25 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.181 |
-0.162 |
| 26 |
248656_at |
AT5G48460
|
[Actin binding Calponin homology (CH) domain-containing protein] |
0.181 |
0.323 |
| 27 |
254391_at |
AT4G21590
|
ENDO3, endonuclease 3 |
0.171 |
-0.222 |
| 28 |
259851_at |
AT1G72250
|
[Di-glucose binding protein with Kinesin motor domain] |
0.170 |
0.434 |
| 29 |
247603_at |
AT5G60930
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.169 |
0.832 |
| 30 |
254807_at |
AT4G12700
|
unknown |
0.166 |
0.680 |
| 31 |
250068_at |
AT5G17960
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.166 |
-1.029 |
| 32 |
263327_at |
AT2G15300
|
[Leucine-rich repeat protein kinase family protein] |
0.163 |
-1.224 |
| 33 |
245861_at |
AT5G28300
|
AtGT2L, GT2L, GT-2Like protein |
0.161 |
0.648 |
| 34 |
263610_at |
AT2G16230
|
[O-Glycosyl hydrolases family 17 protein] |
0.159 |
-0.579 |
| 35 |
257725_at |
AT3G18524
|
ATMSH2, MSH2, MUTS homolog 2 |
0.157 |
0.209 |
| 36 |
248498_at |
AT5G50390
|
EMB3141, EMBRYO DEFECTIVE 3141 |
0.157 |
-0.093 |
| 37 |
249705_at |
AT5G35580
|
[Protein kinase superfamily protein] |
-0.297 |
0.216 |
| 38 |
255064_at |
AT4G08950
|
EXO, EXORDIUM |
-0.277 |
1.130 |
| 39 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
-0.261 |
-0.672 |
| 40 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
-0.220 |
-0.194 |
| 41 |
251749_at |
AT3G55690
|
unknown |
-0.200 |
-0.740 |
| 42 |
248592_at |
AT5G49280
|
[hydroxyproline-rich glycoprotein family protein] |
-0.194 |
-0.020 |
| 43 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
-0.193 |
0.222 |
| 44 |
251806_at |
AT3G55370
|
OBP3, OBF-binding protein 3 |
-0.192 |
0.209 |
| 45 |
260429_at |
AT1G72450
|
JAZ6, jasmonate-zim-domain protein 6, TIFY11B, TIFY DOMAIN PROTEIN 11B |
-0.175 |
0.400 |
| 46 |
251861_at |
AT3G54810
|
BME3, BLUE MICROPYLAR END 3, BME3-ZF, BLUE MICROPYLAR END 3-ZINC FINGER, GATA8, GATA TRANSCRIPTION FACTOR 8 |
-0.172 |
-0.294 |
| 47 |
258025_at |
AT3G19480
|
3-PGDH, PGDH3, phosphoglycerate dehydrogenase 3 |
-0.171 |
-0.240 |
| 48 |
249522_at |
AT5G38700
|
unknown |
-0.149 |
-0.538 |
| 49 |
247878_at |
AT5G57760
|
unknown |
-0.148 |
-1.063 |
| 50 |
262612_at |
AT1G14150
|
PnsL2, Photosynthetic NDH subcomplex L 2, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.143 |
-0.328 |
| 51 |
246453_at |
AT5G16830
|
ATPEP12, ATSYP21, PEP12, PEP12P, SYP21, syntaxin of plants 21 |
-0.139 |
0.444 |
| 52 |
257441_at |
AT2G04020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.136 |
0.207 |
| 53 |
266953_at |
AT2G34540
|
unknown |
-0.132 |
0.250 |
| 54 |
258295_at |
AT3G23400
|
FIB4, fibrillin 4 |
-0.131 |
0.207 |
| 55 |
246030_at |
AT5G21105
|
[Plant L-ascorbate oxidase] |
-0.131 |
0.829 |
| 56 |
253798_at |
AT4G28500
|
ANAC073, NAC domain containing protein 73, NAC073, NAC domain containing protein 73, SND2, SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN 2 |
-0.129 |
-0.248 |
| 57 |
262858_at |
AT1G14940
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.128 |
-1.136 |
| 58 |
256408_at |
AT1G66610
|
[TRAF-like superfamily protein] |
-0.124 |
-0.259 |
| 59 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
-0.121 |
-0.497 |
| 60 |
264852_at |
AT2G17480
|
ATMLO8, MILDEW RESISTANCE LOCUS O 8, MLO8, MILDEW RESISTANCE LOCUS O 8 |
-0.118 |
0.219 |
| 61 |
254794_at |
AT4G12970
|
EPFL9, STOMAGEN, STOMAGEN |
-0.118 |
0.706 |
| 62 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
-0.115 |
0.681 |
| 63 |
265913_at |
AT2G25625
|
unknown |
-0.114 |
-0.739 |
| 64 |
257894_at |
AT3G17100
|
AIF3, ATBS1 Interacting Factor 3 |
-0.110 |
0.452 |
| 65 |
246781_at |
AT5G27350
|
SFP1 |
-0.110 |
-0.234 |
| 66 |
262735_at |
AT1G28630
|
unknown |
-0.108 |
-1.404 |
| 67 |
256799_at |
AT3G18560
|
unknown |
-0.107 |
0.192 |
| 68 |
247461_at |
AT5G62100
|
ATBAG2, BCL-2-associated athanogene 2, BAG2, BCL-2-associated athanogene 2 |
-0.106 |
-1.089 |
| 69 |
259849_at |
AT1G72190
|
[D-isomer specific 2-hydroxyacid dehydrogenase family protein] |
-0.105 |
0.588 |
| 70 |
267305_at |
AT2G30070
|
ATKT1, potassium transporter 1, ATKT1P, ATKUP1, KT1, potassium transporter 1, KUP1, POTASSIUM UPTAKE TRANSPORTER 1 |
-0.101 |
0.635 |
| 71 |
261146_at |
AT1G19620
|
unknown |
-0.100 |
-0.506 |