|
probeID |
AGICode |
Annotation |
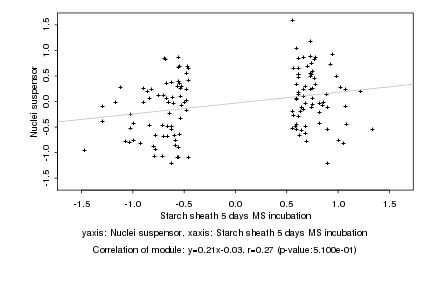
Log2 signal ratio
Starch sheath 5 days MS incubation |
Log2 signal ratio
Nuclei suspensor |
| 1 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
1.332 |
-0.533 |
| 2 |
256787_at |
AT3G13790
|
ATBFRUCT1, ATCWINV1, ARABIDOPSIS THALIANA CELL WALL INVERTASE 1 |
1.211 |
0.209 |
| 3 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
1.081 |
-0.442 |
| 4 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
1.072 |
-0.090 |
| 5 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
1.069 |
0.252 |
| 6 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
1.044 |
-0.813 |
| 7 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
1.019 |
0.290 |
| 8 |
247440_at |
AT5G62680
|
AtNPF2.11, GTR2, GLUCOSINOLATE TRANSPORTER-2, NPF2.11, NRT1/ PTR family 2.11 |
1.004 |
-0.758 |
| 9 |
256825_at |
AT3G22120
|
CWLP, cell wall-plasma membrane linker protein |
0.978 |
0.498 |
| 10 |
253203_at |
AT4G34710
|
ADC2, arginine decarboxylase 2, ATADC2, SPE2 |
0.940 |
0.935 |
| 11 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
0.919 |
0.735 |
| 12 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
0.897 |
-1.204 |
| 13 |
248118_at |
AT5G55050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.895 |
-0.541 |
| 14 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.893 |
-0.116 |
| 15 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.882 |
0.143 |
| 16 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
0.849 |
-0.008 |
| 17 |
252618_at |
AT3G45140
|
ATLOX2, ARABIODOPSIS THALIANA LIPOXYGENASE 2, LOX2, lipoxygenase 2 |
0.842 |
-0.072 |
| 18 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
0.819 |
-0.199 |
| 19 |
256989_at |
AT3G28580
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.816 |
-0.412 |
| 20 |
247607_at |
AT5G60960
|
PNM1, PPR protein localized to the nucleus and mitochondria 1 |
0.811 |
-0.038 |
| 21 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.776 |
0.878 |
| 22 |
259757_at |
AT1G77510
|
ATPDI6, PROTEIN DISULFIDE ISOMERASE 6, ATPDIL1-2, PDI-like 1-2, PDI6, PROTEIN DISULFIDE ISOMERASE 6, PDIL1-2, PDI-like 1-2 |
0.774 |
0.348 |
| 23 |
267181_at |
AT2G37760
|
AKR4C8, Aldo-keto reductase family 4 member C8 |
0.769 |
0.828 |
| 24 |
245145_at |
AT2G45440
|
DHDPS2, dihydrodipicolinate synthase 2 |
0.763 |
0.458 |
| 25 |
259156_at |
AT3G10380
|
ATSEC8, SUBUNIT OF EXOCYST COMPLEX 8, SEC8, subunit of exocyst complex 8 |
0.749 |
0.603 |
| 26 |
255171_at |
AT4G07990
|
[Chaperone DnaJ-domain superfamily protein] |
0.747 |
0.073 |
| 27 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.742 |
0.259 |
| 28 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.742 |
-0.052 |
| 29 |
247164_at |
AT5G65720
|
ATNFS1, ARABIDOPSIS THALIANA NITROGEN FIXATION S (NIFS)-LIKE 1, ATNIFS1, NITROGEN FIXATION S HOMOLOG 1, NFS1, nitrogen fixation S (NIFS)-like 1, NIFS1, NITROGEN FIXATION S HOMOLOG 1 |
0.739 |
-0.105 |
| 30 |
265188_at |
AT1G23800
|
ALDH2B, aldehyde dehydrogenase 2B, ALDH2B7, aldehyde dehydrogenase 2B7 |
0.736 |
0.748 |
| 31 |
262880_at |
AT1G64880
|
[Ribosomal protein S5 family protein] |
0.732 |
0.539 |
| 32 |
262947_at |
AT1G75750
|
GASA1, GAST1 protein homolog 1 |
0.730 |
1.175 |
| 33 |
254649_at |
AT4G18570
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.729 |
0.890 |
| 34 |
251899_at |
AT3G54400
|
[Eukaryotic aspartyl protease family protein] |
0.725 |
0.559 |
| 35 |
259383_at |
AT3G16470
|
JAL35, jacalin-related lectin 35, JR1, JASMONATE RESPONSIVE 1 |
0.724 |
0.238 |
| 36 |
256794_at |
AT3G22230
|
[Ribosomal L27e protein family] |
0.722 |
0.506 |
| 37 |
256983_at |
AT3G13470
|
Cpn60beta2, chaperonin-60beta2 |
0.701 |
0.688 |
| 38 |
245357_at |
AT4G17560
|
[Ribosomal protein L19 family protein] |
0.689 |
-0.775 |
| 39 |
267591_at |
AT2G39705
|
DVL11, DEVIL 11, RTFL8, ROTUNDIFOLIA like 8 |
0.680 |
0.305 |
| 40 |
267367_at |
AT2G44210
|
unknown |
0.680 |
-0.024 |
| 41 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.678 |
-0.481 |
| 42 |
247593_at |
AT5G60790
|
ABCF1, ATP-binding cassette F1, ATGCN1, ARABIDOPSIS THALIANA GENERAL CONTROL NON-REPRESSIBLE 1, GCN1, GENERAL CONTROL NON-REPRESSIBLE 1 |
0.674 |
-0.614 |
| 43 |
248335_at |
AT5G52450
|
[MATE efflux family protein] |
0.663 |
-0.151 |
| 44 |
266256_at |
AT2G27710
|
[60S acidic ribosomal protein family] |
0.661 |
0.868 |
| 45 |
247032_at |
AT5G67240
|
SDN3, small RNA degrading nuclease 3 |
0.658 |
0.116 |
| 46 |
259196_at |
AT3G03600
|
RPS2, ribosomal protein S2 |
0.655 |
0.238 |
| 47 |
251476_at |
AT3G59670
|
unknown |
0.639 |
-0.106 |
| 48 |
245098_at |
AT2G40940
|
ERS, ETHYLENE RESPONSE SENSOR, ERS1, ethylene response sensor 1 |
0.635 |
-0.554 |
| 49 |
265194_at |
AT1G05010
|
ACO4, ethylene forming enzyme, EAT1, EFE, ethylene-forming enzyme |
0.625 |
-0.189 |
| 50 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
0.619 |
-0.650 |
| 51 |
266168_at |
AT2G38870
|
[Serine protease inhibitor, potato inhibitor I-type family protein] |
0.614 |
0.191 |
| 52 |
246921_at |
AT5G25080
|
[Sas10/Utp3/C1D family] |
0.612 |
0.157 |
| 53 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
0.612 |
-0.275 |
| 54 |
251847_at |
AT3G54640
|
TRP3, TRYPTOPHAN-REQUIRING 3, TSA1, tryptophan synthase alpha chain |
0.611 |
0.854 |
| 55 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.610 |
0.484 |
| 56 |
254410_at |
AT4G21410
|
CRK29, cysteine-rich RLK (RECEPTOR-like protein kinase) 29 |
0.609 |
0.656 |
| 57 |
253337_at |
AT4G33470
|
ATHDA14, HDA14, histone deacetylase 14 |
0.608 |
0.536 |
| 58 |
247654_at |
AT5G59850
|
[Ribosomal protein S8 family protein] |
0.607 |
0.537 |
| 59 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.594 |
0.060 |
| 60 |
250054_at |
AT5G17860
|
CAX7, calcium exchanger 7 |
0.591 |
0.048 |
| 61 |
250960_at |
AT5G02940
|
unknown |
0.591 |
-0.439 |
| 62 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
0.588 |
0.351 |
| 63 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.588 |
1.052 |
| 64 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
0.587 |
-0.544 |
| 65 |
252032_at |
AT3G52150
|
PSRP2, plastid-specific ribosomal protein 2 |
0.576 |
-0.470 |
| 66 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.561 |
-0.264 |
| 67 |
257538_at |
AT3G16160
|
[Tesmin/TSO1-like CXC domain-containing protein] |
0.560 |
0.648 |
| 68 |
265671_at |
AT2G32060
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
0.556 |
1.605 |
| 69 |
249466_at |
AT5G39740
|
OLI7, OLIGOCELLULA 7, RPL5B, ribosomal protein L5 B |
0.552 |
-0.516 |
| 70 |
254333_at |
AT4G22753
|
ATSMO1-3, SMO1-3, sterol 4-alpha methyl oxidase 1-3 |
0.551 |
-0.190 |
| 71 |
264495_at |
AT1G27380
|
RIC2, ROP-interactive CRIB motif-containing protein 2 |
-1.476 |
-0.947 |
| 72 |
257896_at |
AT3G16920
|
ATCTL2, CTL2, chitinase-like protein 2 |
-1.302 |
-0.372 |
| 73 |
257757_at |
AT3G18660
|
GUX1, glucuronic acid substitution of xylan 1, PGSIP1, plant glycogenin-like starch initiation protein 1 |
-1.297 |
-0.083 |
| 74 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-1.173 |
-0.005 |
| 75 |
248121_at |
AT5G54690
|
GAUT12, galacturonosyltransferase 12, IRX8, IRREGULAR XYLEM 8, LGT6 |
-1.125 |
0.287 |
| 76 |
249070_at |
AT5G44030
|
CESA4, cellulose synthase A4, IRX5, IRREGULAR XYLEM 5, NWS2 |
-1.079 |
-0.774 |
| 77 |
251297_at |
AT3G62020
|
GLP10, germin-like protein 10 |
-1.037 |
-0.801 |
| 78 |
253798_at |
AT4G28500
|
ANAC073, NAC domain containing protein 73, NAC073, NAC domain containing protein 73, SND2, SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN 2 |
-1.029 |
-0.240 |
| 79 |
250933_at |
AT5G03170
|
ATFLA11, ARABIDOPSIS FASCICLIN-LIKE ARABINOGALACTAN-PROTEIN 11, FLA11, FASCICLIN-like arabinogalactan-protein 11 |
-1.027 |
-0.522 |
| 80 |
261928_at |
AT1G22480
|
[Cupredoxin superfamily protein] |
-1.003 |
-0.422 |
| 81 |
257233_at |
AT3G15050
|
IQD10, IQ-domain 10 |
-1.003 |
-0.748 |
| 82 |
246512_at |
AT5G15630
|
COBL4, COBRA-LIKE4, IRX6, IRREGULAR XYLEM 6 |
-0.931 |
-0.817 |
| 83 |
262438_at |
AT1G47410
|
unknown |
-0.901 |
-0.005 |
| 84 |
249227_at |
AT5G42180
|
PER64, peroxidase 64 |
-0.899 |
0.269 |
| 85 |
263942_at |
AT2G35860
|
FLA16, FASCICLIN-like arabinogalactan protein 16 precursor |
-0.860 |
0.201 |
| 86 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
-0.841 |
-0.464 |
| 87 |
250233_at |
AT5G13460
|
IQD11, IQ-domain 11 |
-0.839 |
0.060 |
| 88 |
254618_at |
AT4G18780
|
ATCESA8, CELLULOSE SYNTHASE 8, CESA8, CELLULOSE SYNTHASE 8, IRX1, IRREGULAR XYLEM 1, LEW2, LEAF WILTING 2 |
-0.825 |
0.251 |
| 89 |
260326_at |
AT1G63910
|
AtMYB103, myb domain protein 103, MYB103, myb domain protein 103 |
-0.807 |
-0.869 |
| 90 |
262922_at |
AT1G79420
|
unknown |
-0.798 |
-1.074 |
| 91 |
247812_at |
AT5G58390
|
[Peroxidase superfamily protein] |
-0.786 |
-0.925 |
| 92 |
266708_at |
AT2G03200
|
[Eukaryotic aspartyl protease family protein] |
-0.785 |
-0.660 |
| 93 |
261425_at |
AT1G18880
|
AtNPF2.9, NPF2.9, NRT1/ PTR family 2.9, NRT1.9, nitrate transporter 1.9 |
-0.756 |
0.131 |
| 94 |
253010_at |
AT4G37750
|
ANT, AINTEGUMENTA, CKC, CKC1, COMPLEMENTING A PROTEIN KINASE C MUTANT 1, DRG, DRAGON |
-0.714 |
-0.461 |
| 95 |
262858_at |
AT1G14940
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.714 |
-1.067 |
| 96 |
265545_at |
AT2G28250
|
NCRK |
-0.710 |
-0.668 |
| 97 |
250100_at |
AT5G16570
|
GLN1;4, glutamine synthetase 1;4 |
-0.708 |
0.135 |
| 98 |
260531_at |
AT2G47240
|
CER8, ECERIFERUM 8, LACS1, LONG-CHAIN ACYL-COA SYNTHASE 1 |
-0.692 |
0.845 |
| 99 |
246425_at |
AT5G17420
|
ATCESA7, CESA7, CELLULOSE SYNTHASE CATALYTIC SUBUNIT 7, IRX3, IRREGULAR XYLEM 3, MUR10, MURUS 10 |
-0.684 |
0.835 |
| 100 |
251509_at |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
-0.680 |
0.372 |
| 101 |
256293_at |
AT1G69440
|
AGO7, ARGONAUTE7, ZIP, ZIPPY |
-0.675 |
0.076 |
| 102 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.665 |
-0.485 |
| 103 |
263330_at |
AT2G15320
|
[Leucine-rich repeat (LRR) family protein] |
-0.663 |
-0.677 |
| 104 |
258040_at |
AT3G21190
|
AtMSR1, MSR1, Mannan Synthesis Related 1 |
-0.661 |
-0.011 |
| 105 |
253501_at |
AT4G32010
|
HSI2-L1, HSL1, HSI2-like 1, VAL2, VP1/ABI3-LIKE 2 |
-0.660 |
-0.013 |
| 106 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
-0.646 |
-0.220 |
| 107 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
-0.629 |
0.374 |
| 108 |
247638_at |
AT5G60490
|
AtFLA12, FLA12, FASCICLIN-like arabinogalactan-protein 12 |
-0.627 |
-0.481 |
| 109 |
257467_at |
AT1G31320
|
LBD4, LOB domain-containing protein 4 |
-0.626 |
-1.194 |
| 110 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
-0.625 |
-0.536 |
| 111 |
247663_at |
AT5G60110
|
APUM18, pumilio 18, PUM18, pumilio 18 |
-0.620 |
0.089 |
| 112 |
256926_at |
AT3G22540
|
unknown |
-0.604 |
-0.030 |
| 113 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
-0.603 |
-0.652 |
| 114 |
264559_at |
AT1G09610
|
GXM3, glucuronoxylan methyltransferase 3 |
-0.590 |
-0.852 |
| 115 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
-0.587 |
-0.759 |
| 116 |
253113_at |
AT4G35985
|
[Senescence/dehydration-associated protein-related] |
-0.574 |
0.301 |
| 117 |
256435_at |
AT3G11180
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.568 |
-1.085 |
| 118 |
255246_at |
AT4G05640
|
unknown |
-0.561 |
-0.892 |
| 119 |
247648_at |
AT5G60020
|
ATLAC17, LAC17, laccase 17 |
-0.560 |
0.877 |
| 120 |
260004_at |
AT1G67860
|
unknown |
-0.557 |
0.402 |
| 121 |
246487_at |
AT5G16030
|
unknown |
-0.556 |
0.677 |
| 122 |
248355_at |
AT5G52340
|
ATEXO70A2, exocyst subunit exo70 family protein A2, EXO70A2, exocyst subunit exo70 family protein A2 |
-0.556 |
-1.083 |
| 123 |
256673_at |
AT3G52370
|
FLA15, FASCICLIN-like arabinogalactan protein 15 precursor |
-0.550 |
0.697 |
| 124 |
253193_at |
AT4G35380
|
[SEC7-like guanine nucleotide exchange family protein] |
-0.549 |
-0.633 |
| 125 |
245386_at |
AT4G14010
|
RALFL32, ralf-like 32 |
-0.547 |
0.360 |
| 126 |
245159_at |
AT2G33100
|
ATCSLD1, cellulose synthase-like D1, CSLD1, cellulose synthase-like D1, CSLD1, CELLULOSE-SYNTHASE LIKE D1 |
-0.543 |
0.256 |
| 127 |
262060_at |
AT1G80170
|
[Pectin lyase-like superfamily protein] |
-0.541 |
0.107 |
| 128 |
254753_at |
AT4G13190
|
[Protein kinase superfamily protein] |
-0.536 |
-0.323 |
| 129 |
250653_at |
AT5G06930
|
[LOCATED IN: chloroplast] |
-0.530 |
-0.066 |
| 130 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
-0.529 |
0.304 |
| 131 |
250216_at |
AT5G14090
|
AtLAZY1, LAZY1, LAZY 1 |
-0.500 |
-0.008 |
| 132 |
257690_at |
AT3G12830
|
SAUR72, SMALL AUXIN UPREGULATED 72 |
-0.486 |
0.031 |
| 133 |
251448_at |
AT3G59845
|
[Zinc-binding dehydrogenase family protein] |
-0.485 |
0.554 |
| 134 |
247522_at |
AT5G61340
|
unknown |
-0.481 |
0.245 |
| 135 |
265234_at |
AT2G07721
|
unknown |
-0.479 |
-0.173 |
| 136 |
253580_at |
AT4G30400
|
[RING/U-box superfamily protein] |
-0.469 |
0.704 |
| 137 |
261046_at |
AT1G01390
|
[UDP-Glycosyltransferase superfamily protein] |
-0.467 |
-1.095 |
| 138 |
263689_at |
AT1G26820
|
RNS3, ribonuclease 3 |
-0.466 |
0.661 |
| 139 |
265754_x_at |
AT2G10640
|
[CACTA-like transposase family (Ptta/En/Spm), has a 7.4e-42 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.462 |
0.414 |